Label-Free Relative Quantitation of Isobaric and Isomeric Human Histone H2A and H2B Variants by Fourier Transform Ion Cyclotron Resonance Top-Down MS/MS
- PMID: 27431976
- PMCID: PMC6261780
- DOI: 10.1021/acs.jproteome.6b00414
Label-Free Relative Quantitation of Isobaric and Isomeric Human Histone H2A and H2B Variants by Fourier Transform Ion Cyclotron Resonance Top-Down MS/MS
Abstract
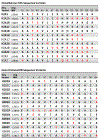
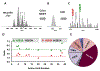
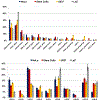
Histone variants are known to play a central role in genome regulation and maintenance. However, many variants are inaccessible by antibody-based methods or bottom-up tandem mass spectrometry due to their highly similar sequences. For many, the only tractable approach is with intact protein top-down tandem mass spectrometry. Here, ultra-high-resolution FT-ICR MS and MS/MS yield quantitative relative abundances of all detected HeLa H2A and H2B isobaric and isomeric variants with a label-free approach. We extend the analysis to identify and relatively quantitate 16 proteoforms from 12 sequence variants of histone H2A and 10 proteoforms of histone H2B from three other cell lines: human embryonic stem cells (WA09), U937, and a prostate cancer cell line LaZ. The top-down MS/MS approach provides a path forward for more extensive elucidation of the biological role of many previously unstudied histone variants and post-translational modifications.
Keywords: FT-ICR; FTMS; histone; post-translational modification; proteoform; sequence variants.
Figures


Similar articles
-
Fourier-transform ion cyclotron resonance mass spectrometry for characterizing proteoforms.Mass Spectrom Rev. 2022 Mar;41(2):158-177. doi: 10.1002/mas.21653. Epub 2020 Sep 7. Mass Spectrom Rev. 2022. PMID: 32894796 Free PMC article. Review.
-
Histone Analysis Using Mobility- and Mass-Selected Ultraviolet Photodissociation in Tandem with Ultra-High-Resolution Mass Spectrometry.Anal Chem. 2025 Jul 8;97(26):13965-13973. doi: 10.1021/acs.analchem.5c02166. Epub 2025 Jun 24. Anal Chem. 2025. PMID: 40552939 Free PMC article.
-
A Top-Down Proteomics Platform Coupling Serial Size Exclusion Chromatography and Fourier Transform Ion Cyclotron Resonance Mass Spectrometry.Anal Chem. 2019 Mar 19;91(6):3835-3844. doi: 10.1021/acs.analchem.8b04082. Epub 2019 Feb 25. Anal Chem. 2019. PMID: 30758949 Free PMC article.
-
One-Pot Quantitative Top- and Middle-Down Analysis of GluC-Digested Histone H4.J Am Soc Mass Spectrom. 2019 Dec;30(12):2514-2525. doi: 10.1007/s13361-019-02219-1. Epub 2019 May 30. J Am Soc Mass Spectrom. 2019. PMID: 31147891
-
Towards analytically useful two-dimensional Fourier transform ion cyclotron resonance mass spectrometry.Anal Bioanal Chem. 2013 Jan;405(1):51-61. doi: 10.1007/s00216-012-6422-8. Epub 2012 Oct 18. Anal Bioanal Chem. 2013. PMID: 23076397 Review.
Cited by
-
Histone proteoform analysis reveals epigenetic changes in adult mouse brown adipose tissue in response to cold stress.bioRxiv [Preprint]. 2024 Jan 22:2023.07.30.551059. doi: 10.1101/2023.07.30.551059. bioRxiv. 2024. Update in: Epigenetics Chromatin. 2024 Apr 27;17(1):12. doi: 10.1186/s13072-024-00536-8. PMID: 38328142 Free PMC article. Updated. Preprint.
-
Bullet points to evaluate the performance of the middle-down proteomics workflow for histone modification analysis.Methods. 2020 Dec 1;184:86-92. doi: 10.1016/j.ymeth.2020.01.013. Epub 2020 Feb 15. Methods. 2020. PMID: 32070774 Free PMC article.
-
Direct histone proteoform profiling of the unannotated, endangered coral Acropora cervicornis.Nucleic Acids Res. 2025 Jul 19;53(14):gkaf740. doi: 10.1093/nar/gkaf740. Nucleic Acids Res. 2025. PMID: 40744498 Free PMC article.
-
Bottom-up Histone Post-translational Modification Analysis using Liquid Chromatography, Trapped Ion Mobility Spectrometry, and Tandem Mass Spectrometry.J Proteome Res. 2024 Sep 6;23(9):3867-3876. doi: 10.1021/acs.jproteome.4c00177. Epub 2024 Aug 23. J Proteome Res. 2024. PMID: 39177337
-
High-Throughput Quantitative Top-Down Proteomics: Histone H4.J Am Soc Mass Spectrom. 2019 Dec;30(12):2548-2560. doi: 10.1007/s13361-019-02350-z. Epub 2019 Nov 18. J Am Soc Mass Spectrom. 2019. PMID: 31741267
References
-
- Biel M; Wascholowski V; Giannis A Epigenetics-an epicenter of gene regulation: histones and histone-modifying enzymes. Angew. Chem. Int. Edit 2005, 44, 3186–3216. - PubMed
-
- Reik W Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 2007, 447, 425–432. - PubMed
-
- Liu C; Liu L; Shan J; Shen J; Xu Y; Zhang Q; Yang Z; Wu L; Xia F; Bie P; Cui Y; Zhang X; Bian X; Quan C Histone deacetylase 3 participates in self-renewal of liver cancer stem cells through histone modification. Cancer Letters 2013, 339, 60–69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

