Genome-Wide Meta-Analyses of Breast, Ovarian, and Prostate Cancer Association Studies Identify Multiple New Susceptibility Loci Shared by at Least Two Cancer Types
- PMID: 27432226
- PMCID: PMC5010513
- DOI: 10.1158/2159-8290.CD-15-1227
Genome-Wide Meta-Analyses of Breast, Ovarian, and Prostate Cancer Association Studies Identify Multiple New Susceptibility Loci Shared by at Least Two Cancer Types
Abstract
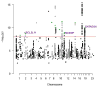
Breast, ovarian, and prostate cancers are hormone-related and may have a shared genetic basis, but this has not been investigated systematically by genome-wide association (GWA) studies. Meta-analyses combining the largest GWA meta-analysis data sets for these cancers totaling 112,349 cases and 116,421 controls of European ancestry, all together and in pairs, identified at P < 10(-8) seven new cross-cancer loci: three associated with susceptibility to all three cancers (rs17041869/2q13/BCL2L11; rs7937840/11q12/INCENP; rs1469713/19p13/GATAD2A), two breast and ovarian cancer risk loci (rs200182588/9q31/SMC2; rs8037137/15q26/RCCD1), and two breast and prostate cancer risk loci (rs5013329/1p34/NSUN4; rs9375701/6q23/L3MBTL3). Index variants in five additional regions previously associated with only one cancer also showed clear association with a second cancer type. Cell-type-specific expression quantitative trait locus and enhancer-gene interaction annotations suggested target genes with potential cross-cancer roles at the new loci. Pathway analysis revealed significant enrichment of death receptor signaling genes near loci with P < 10(-5) in the three-cancer meta-analysis.
Significance: We demonstrate that combining large-scale GWA meta-analysis findings across cancer types can identify completely new risk loci common to breast, ovarian, and prostate cancers. We show that the identification of such cross-cancer risk loci has the potential to shed new light on the shared biology underlying these hormone-related cancers. Cancer Discov; 6(9); 1052-67. ©2016 AACR.This article is highlighted in the In This Issue feature, p. 932.
©2016 American Association for Cancer Research.
Conflict of interest statement
The authors disclose no potential conflicts of interest.
Figures



Similar articles
-
Bromodomain protein 4 discriminates tissue-specific super-enhancers containing disease-specific susceptibility loci in prostate and breast cancer.BMC Genomics. 2017 Mar 31;18(1):270. doi: 10.1186/s12864-017-3620-y. BMC Genomics. 2017. PMID: 28359301 Free PMC article.
-
Identification of independent association signals and putative functional variants for breast cancer risk through fine-scale mapping of the 12p11 locus.Breast Cancer Res. 2016 Jun 21;18(1):64. doi: 10.1186/s13058-016-0718-0. Breast Cancer Res. 2016. PMID: 27459855 Free PMC article.
-
Multiple loci with different cancer specificities within the 8q24 gene desert.J Natl Cancer Inst. 2008 Jul 2;100(13):962-6. doi: 10.1093/jnci/djn190. Epub 2008 Jun 24. J Natl Cancer Inst. 2008. PMID: 18577746 Free PMC article.
-
Common Genetic Variation and Susceptibility to Ovarian Cancer: Current Insights and Future Directions.Cancer Epidemiol Biomarkers Prev. 2018 Apr;27(4):395-404. doi: 10.1158/1055-9965.EPI-17-0315. Epub 2017 Jun 14. Cancer Epidemiol Biomarkers Prev. 2018. PMID: 28615364 Review.
-
A compendium of genome-wide associations for cancer: critical synopsis and reappraisal.J Natl Cancer Inst. 2010 Jun 16;102(12):846-58. doi: 10.1093/jnci/djq173. Epub 2010 May 26. J Natl Cancer Inst. 2010. PMID: 20505153 Free PMC article. Review.
Cited by
-
Screening and Identification of Therapeutic Targets for Pulmonary Arterial Hypertension Through Microarray Technology.Front Genet. 2020 Jul 22;11:782. doi: 10.3389/fgene.2020.00782. eCollection 2020. Front Genet. 2020. PMID: 32849793 Free PMC article.
-
Identifying disease-associated copy number variations by a doubly penalized regression model.Biometrics. 2018 Dec;74(4):1341-1350. doi: 10.1111/biom.12920. Epub 2018 Jun 12. Biometrics. 2018. PMID: 29894562 Free PMC article.
-
Exploring Shared Susceptibility between Two Neural Crest Cells Originating Conditions: Neuroblastoma and Congenital Heart Disease.Genes (Basel). 2019 Aug 30;10(9):663. doi: 10.3390/genes10090663. Genes (Basel). 2019. PMID: 31480262 Free PMC article.
-
Mapping inherited genetic variation with opposite effects on autoimmune disease and cancer identifies candidate drug targets associated with the anti-tumor immune response.medRxiv [Preprint]. 2023 Dec 28:2023.12.23.23300491. doi: 10.1101/2023.12.23.23300491. medRxiv. 2023. Update in: Genes (Basel). 2025 May 14;16(5):575. doi: 10.3390/genes16050575. PMID: 38234717 Free PMC article. Updated. Preprint.
-
Cancer risk susceptibility loci in a Swedish population.Oncotarget. 2017 Nov 25;8(66):110300-110310. doi: 10.18632/oncotarget.22687. eCollection 2017 Dec 15. Oncotarget. 2017. PMID: 29299148 Free PMC article.
References
-
- Henderson BE, Feigelson HS. Hormonal carcinogenesis. Carcinogenesis. 2000;21:427–33. - PubMed
-
- U.S. Cancer Statistics Working Group. United States Cancer Statistics: 1999–2012 Incidence and Mortality Web-based Report. [Internet] Atlanta: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute; 2015. [cited 2015 Sep 22]. Available from: www.cdc.gov/uscs.
-
- Sellers TA, Potter JD, Rich SS, Drinkard CR, Bostick RM, Kushi LH, et al. Familial clustering of breast and prostate cancers and risk of postmenopausal breast cancer. J Natl Cancer Inst. 1994;86:1860–5. - PubMed
-
- Tung K-H, Goodman MT, Wu AH, McDuffie K, Wilkens LR, Nomura AMY, et al. Aggregation of ovarian cancer with breast, ovarian, colorectal, and prostate cancer in first-degree relatives. Am J Epidemiol. 2004;159:750–8. - PubMed
-
- Cerhan JR, Parker AS, Putnam SD, Chiu BC, Lynch CF, Cohen MB, et al. Family history and prostate cancer risk in a population-based cohort of Iowa men. Cancer Epidemiol Biomarkers Prev. 1999;8:53–60. - PubMed
Publication types
MeSH terms
Grants and funding
- 16565/CRUK_/Cancer Research UK/United Kingdom
- R01 CA092447/CA/NCI NIH HHS/United States
- C12292/A11174/CRUK_/Cancer Research UK/United Kingdom
- G1000143/MRC_/Medical Research Council/United Kingdom
- C5047/A3354/CRUK_/Cancer Research UK/United Kingdom
- G0401527/MRC_/Medical Research Council/United Kingdom
- C1281/A12014/CRUK_/Cancer Research UK/United Kingdom
- G0501974/MRC_/Medical Research Council/United Kingdom
- 16491/CRUK_/Cancer Research UK/United Kingdom
- C536/A13086/CRUK_/Cancer Research UK/United Kingdom
- R01 CA176785/CA/NCI NIH HHS/United States
- 17528/CRUK_/Cancer Research UK/United Kingdom
- C490/A6187/CRUK_/Cancer Research UK/United Kingdom
- C1287/A12014 /CRUK_/Cancer Research UK/United Kingdom
- C5047/A7357/CRUK_/Cancer Research UK/United Kingdom
- C1287/A10710/CRUK_/Cancer Research UK/United Kingdom
- MR/N003284/1/MRC_/Medical Research Council/United Kingdom
- P30 CA046592/CA/NCI NIH HHS/United States
- C16913/A6135/CRUK_/Cancer Research UK/United Kingdom
- C490/A10119/CRUK_/Cancer Research UK/United Kingdom
- C536/A6689/CRUK_/Cancer Research UK/United Kingdom
- UL1 TR001863/TR/NCATS NIH HHS/United States
- C5047/A10692/CRUK_/Cancer Research UK/United Kingdom
- R00 CA184415/CA/NCI NIH HHS/United States
- C490/A10124/CRUK_/Cancer Research UK/United Kingdom
- U01 CA116167/CA/NCI NIH HHS/United States
- C5047/A8384/CRUK_/Cancer Research UK/United Kingdom
- P30 CA008748/CA/NCI NIH HHS/United States
- R01 CA128978/CA/NCI NIH HHS/United States
- 14136/CRUK_/Cancer Research UK/United Kingdom
- U19 CA148537/CA/NCI NIH HHS/United States
- C5047/A15007/CRUK_/Cancer Research UK/United Kingdom
- 15007/CRUK_/Cancer Research UK/United Kingdom
- 19170/CRUK_/Cancer Research UK/United Kingdom
- U19 CA148112/CA/NCI NIH HHS/United States
- U19 CA148065/CA/NCI NIH HHS/United States
- 10119/CRUK_/Cancer Research UK/United Kingdom
- C8197/A16565/CRUK_/Cancer Research UK/United Kingdom
- 10118/CRUK_/Cancer Research UK/United Kingdom
- C1287/A10118/CRUK_/Cancer Research UK/United Kingdom
- 001/WHO_/World Health Organization/International
- P50 CA136393/CA/NCI NIH HHS/United States
- C1287/A10118 /CRUK_/Cancer Research UK/United Kingdom
- P20 GM104416/GM/NIGMS NIH HHS/United States
- 10124/CRUK_/Cancer Research UK/United Kingdom
- 16561/CRUK_/Cancer Research UK/United Kingdom
- 13065/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

