Application of network construction to estimate functional changes to insulin receptor substrates 1 and 2 in Huh7 cells following infection with the hepatitis C virus
- PMID: 27432476
- PMCID: PMC4991679
- DOI: 10.3892/mmr.2016.5527
Application of network construction to estimate functional changes to insulin receptor substrates 1 and 2 in Huh7 cells following infection with the hepatitis C virus
Abstract
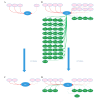
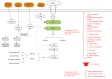
Hepatitis C virus (HCV) is closely associated with insulin resistance (IS), acting primarily by interfering with insulin signaling pathways, increasing cytokine-mediated (tumor necrosis factor α, interleukin 6) inflammatory responses and enhancing oxidative stress. In the insulin signaling pathways, the insulin receptor substrate (IRS) is one of the key regulatory factors. The present study constructed gene regulatory sub‑networks specific for IRS1 and IRS2 in Huh7 cells and HCV‑infected Huh7 (HCV‑Huh7) cells using linear programming and a decomposition algorithm, and investigated the possible mechanisms underlying the function of IRS1/2 in HCV‑induced IS in Huh7 cells. All data were obtained from GSE20948 of the Gene Expression Omnibus database from the National Center for Biotechnology Information. Genes which were significantly differentially expressed between Huh7 and HCV‑Huh7 cells were analyzed using the significance analysis of microarray algorithm. The top 50 genes, including IRS1/2, were used as target genes to determine the gene regulatory networks and next the sub‑networks of IRS1 and IRS2 in HCV‑Huh7 and Huh7 cells using Gene Regulatory Network Inference Tool, an algorithm based on linear programming and the decomposition process. The IRS1/2 sub‑networks were divided into upstream/downstream groups and activation/suppression clusters, and were further analyzed using Molecule Annotation System 3.0 and Database for Annotation, Visualization, and Integrated Discovery software, two online platforms for enrichment and clustering analysis and visualization. The results indicated that in Huh7 cells, the downstream network of IRS2 is more complex than that of IRS1, indicating that the insulin metabolism in Huh7 cells may be primarily mediated by IRS2. In HCV‑Huh7 cells, the downstream pathway of IRS2 is blocked, suggesting that this may be the underlying mechanism in HCV infection that leads to insulin resistance. The present findings add a further dimension to the understanding of the pathological mechanisms of HCV infection-associated insulin resistance, and provide novel concepts for insulin resistance and glucose metabolism research.
Figures
Similar articles
-
Computational networks of activating transcription factor 3 gene in Huh7 cell lines and hepatitis C virus-infected Huh7 cell lines.Mol Med Rep. 2015 Jul;12(1):1239-46. doi: 10.3892/mmr.2015.3548. Epub 2015 Mar 26. Mol Med Rep. 2015. PMID: 25816118
-
Hepatitis C virus down-regulates insulin receptor substrates 1 and 2 through up-regulation of suppressor of cytokine signaling 3.Am J Pathol. 2004 Nov;165(5):1499-508. doi: 10.1016/S0002-9440(10)63408-6. Am J Pathol. 2004. PMID: 15509521 Free PMC article.
-
Inhibition of IRS-1 by hepatitis C virus infection leads to insulin resistance in a PTEN-dependent manner.Virol J. 2015 Feb 3;12:12. doi: 10.1186/s12985-015-0241-4. Virol J. 2015. PMID: 25645159 Free PMC article.
-
Selective Insulin Resistance in the Kidney.Biomed Res Int. 2016;2016:5825170. doi: 10.1155/2016/5825170. Epub 2016 May 9. Biomed Res Int. 2016. PMID: 27247938 Free PMC article. Review.
-
Divergent Roles of IRS (Insulin Receptor Substrate) 1 and 2 in Liver and Skeletal Muscle.Curr Med Chem. 2017;24(17):1827-1852. doi: 10.2174/0929867324666170426142826. Curr Med Chem. 2017. PMID: 28462703 Review.
Cited by
-
Characterization of Huh7 cells after the induction of insulin resistance and post-treatment with metformin.Cytotechnology. 2020 Aug;72(4):499-511. doi: 10.1007/s10616-020-00398-4. Epub 2020 May 14. Cytotechnology. 2020. PMID: 32409919 Free PMC article.
-
Effects of Coatings on Antioxidant Enzyme Activities, Histopathology, and Transcriptome Profiles of Kidney Tissue in Larimichthys crocea.Genes (Basel). 2025 Mar 29;16(4):392. doi: 10.3390/genes16040392. Genes (Basel). 2025. PMID: 40282352 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

