RNA polymerase II promoter-proximal pausing in mammalian long non-coding genes
- PMID: 27432546
- PMCID: PMC6524647
- DOI: 10.1016/j.ygeno.2016.07.003
RNA polymerase II promoter-proximal pausing in mammalian long non-coding genes
Abstract
Mammalian genomes encode a large number of non-coding RNAs (ncRNAs) that greatly exceed mRNA genes. While the physiological and pathological roles of ncRNAs have been increasingly understood, the mechanisms of regulation of ncRNA expression are less clear. Here, our genomic study has shown that a significant number of long non-coding RNAs (lncRNAs, >1000 nucleotides) harbor RNA polymerase II (Pol II) engaged with the transcriptional start site. A pausing and transcriptional elongation factor for protein-coding genes, tripartite motif-containing 28 (TRIM28) regulates the transcription of a subset of lncRNAs in mammalian cells. In addition, the majority of lncRNAs in human and murine cells regulated by Pol II promoter-proximal pausing appear to function in stimulus-inducible biological pathways. Our findings suggest an important role of Pol II pausing for the transcription of mammalian lncRNA genes.
Keywords: Long non-coding RNAs; RNA polymerase II promoter-proximal pausing; TRIM28.
Copyright © 2016 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures
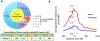
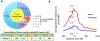
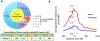
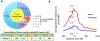
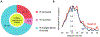
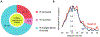
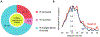
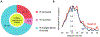





References
-
- Bierhoff H, Dammert MA, Brocks D, Dambacher S, Schotta G, Grummt I: Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol Cell 2014, 54:675–682. - PubMed
-
- Lee JT, Bartolomei MS: X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 2013, 152:1308–1323. - PubMed
-
- Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL: Long noncoding RNAs with snoRNA ends. Mol Cell 2012, 48:219–230. - PubMed
-
- Carpenter S, Ricci EP, Mercier BC, Moore MJ, Fitzgerald KA: Post-transcriptional regulation of gene expression in innate immunity. Nat Rev Immunol 2014, 14:361–376. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

