The novel 2016 WHO Neisseria gonorrhoeae reference strains for global quality assurance of laboratory investigations: phenotypic, genetic and reference genome characterization
- PMID: 27432602
- PMCID: PMC5079299
- DOI: 10.1093/jac/dkw288
The novel 2016 WHO Neisseria gonorrhoeae reference strains for global quality assurance of laboratory investigations: phenotypic, genetic and reference genome characterization
Abstract
Objectives: Gonorrhoea and MDR Neisseria gonorrhoeae remain public health concerns globally. Enhanced, quality-assured, gonococcal antimicrobial resistance (AMR) surveillance is essential worldwide. The WHO global Gonococcal Antimicrobial Surveillance Programme (GASP) was relaunched in 2009. We describe the phenotypic, genetic and reference genome characteristics of the 2016 WHO gonococcal reference strains intended for quality assurance in the WHO global GASP, other GASPs, diagnostics and research worldwide.
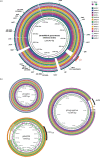
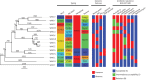
Methods: The 2016 WHO reference strains (n = 14) constitute the eight 2008 WHO reference strains and six novel strains. The novel strains represent low-level to high-level cephalosporin resistance, high-level azithromycin resistance and a porA mutant. All strains were comprehensively characterized for antibiogram (n = 23), serovar, prolyliminopeptidase, plasmid types, molecular AMR determinants, N. gonorrhoeae multiantigen sequence typing STs and MLST STs. Complete reference genomes were produced using single-molecule PacBio sequencing.
Results: The reference strains represented all available phenotypes, susceptible and resistant, to antimicrobials previously and currently used or considered for future use in gonorrhoea treatment. All corresponding resistance genotypes and molecular epidemiological types were described. Fully characterized, annotated and finished references genomes (n = 14) were presented.
Conclusions: The 2016 WHO gonococcal reference strains are intended for internal and external quality assurance and quality control in laboratory investigations, particularly in the WHO global GASP and other GASPs, but also in phenotypic (e.g. culture, species determination) and molecular diagnostics, molecular AMR detection, molecular epidemiology and as fully characterized, annotated and finished reference genomes in WGS analysis, transcriptomics, proteomics and other molecular technologies and data analysis.
© The Author 2016. Published by Oxford University Press on behalf of the British Society for Antimicrobial Chemotherapy. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures
References
-
- WHO. Global Action Plan to Control the Spread and Impact of Antimicrobial Resistance in Neisseria gonorrhoeae. 2012. http://whqlibdoc.who.int/publications/2012/9789241503501_eng.pdf?ua=1.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

