Coordinate Regulation of Metabolite Glycosylation and Stress Hormone Biosynthesis by TT8 in Arabidopsis
- PMID: 27432888
- PMCID: PMC4972274
- DOI: 10.1104/pp.16.00421
Coordinate Regulation of Metabolite Glycosylation and Stress Hormone Biosynthesis by TT8 in Arabidopsis
Abstract
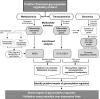
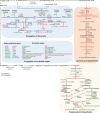
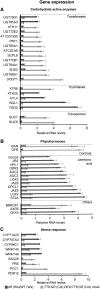
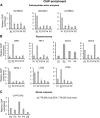
Secondary metabolites play a key role in coordinating ecology and defense strategies of plants. Diversity of these metabolites arises by conjugation of core structures with diverse chemical moieties, such as sugars in glycosylation. Active pools of phytohormones, including those involved in plant stress response, are also regulated by glycosylation. While much is known about the enzymes involved in glycosylation, we know little about their regulation or coordination with other processes. We characterized the flavonoid pathway transcription factor TRANSPARENT TESTA8 (TT8) in Arabidopsis (Arabidopsis thaliana) using an integrative omics strategy. This approach provides a systems-level understanding of the cellular machinery that is used to generate metabolite diversity by glycosylation. Metabolomics analysis of TT8 loss-of-function and inducible overexpression lines showed that TT8 coordinates glycosylation of not only flavonoids, but also nucleotides, thus implicating TT8 in regulating pools of activated nucleotide sugars. Transcriptome and promoter network analyses revealed that the TT8 regulome included sugar transporters, proteins involved in sugar binding and sequestration, and a number of carbohydrate-active enzymes. Importantly, TT8 affects stress response, along with brassinosteroid and jasmonic acid biosynthesis, by directly binding to the promoters of key genes of these processes. This combined effect on metabolite glycosylation and stress hormones by TT8 inducible overexpression led to significant increase in tolerance toward multiple abiotic and biotic stresses. Conversely, loss of TT8 leads to increased sensitivity to these stresses. Thus, the transcription factor TT8 is an integrator of secondary metabolism and stress response. These findings provide novel approaches to improve broad-spectrum stress tolerance.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures




References
-
- Bajguz A. (2007) Metabolism of brassinosteroids in plants. Plant Physiol Biochem 45: 95–107 - PubMed
-
- Bajguz A, Piotrowska A (2009) Conjugates of auxin and cytokinin. Phytochemistry 70: 957–969 - PubMed
-
- Bari R, Jones JD (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488 - PubMed
-
- Baudry A, Caboche M, Lepiniec L (2006) TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J 46: 768–779 - PubMed
-
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39: 366–380 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

