Mutations in STN1 cause Coats plus syndrome and are associated with genomic and telomere defects
- PMID: 27432940
- PMCID: PMC4986528
- DOI: 10.1084/jem.20151618
Mutations in STN1 cause Coats plus syndrome and are associated with genomic and telomere defects
Abstract
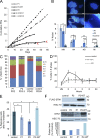
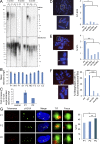
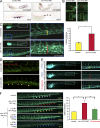
The analysis of individuals with telomere defects may shed light on the delicate interplay of factors controlling genome stability, premature aging, and cancer. We herein describe two Coats plus patients with telomere and genomic defects; both harbor distinct, novel mutations in STN1, a member of the human CTC1-STN1-TEN1 (CST) complex, thus linking this gene for the first time to a human telomeropathy. We characterized the patients' phenotype, recapitulated it in a zebrafish model and rescued cellular and clinical aspects by the ectopic expression of wild-type STN1 or by thalidomide treatment. Interestingly, a significant lengthy control of the gastrointestinal bleeding in one of our patients was achieved by thalidomide treatment, exemplifying a successful bed-to-bench-and-back approach.
© 2016 Simon et al.
Figures

References
-
- Anderson B.H., Kasher P.R., Mayer J., Szynkiewicz M., Jenkinson E.M., Bhaskar S.S., Urquhart J.E., Daly S.B., Dickerson J.E., O’Sullivan J., et al. . 2012. Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat. Genet. 44:338–342. 10.1038/ng.1084 - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

