TNF activation of NF-κB is essential for development of single-positive thymocytes
- PMID: 27432943
- PMCID: PMC4986527
- DOI: 10.1084/jem.20151604
TNF activation of NF-κB is essential for development of single-positive thymocytes
Abstract
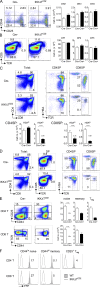
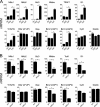
NF-κB activation has been implicated at multiple stages of thymic development of T cells, during which it is thought to mediate developmental signals originating from the T cell receptor (TCR). However, the Card11-Bcl10-Malt1 (CBM) complex that is essential for TCR activation of NF-κB in peripheral T cells is not required for thymocyte development. It has remained unclear whether the TCR activates NF-κB independent of the CBM complex in thymocyte development or whether another NF-κB activating receptor is involved. In the present study, we generated mice in which T cells lacked expression of both catalytic subunits of the inhibitor of κB kinase (IKK) complex, IKK1 and IKK2, to investigate this question. Although early stages of T cell development were unperturbed, maturation of CD4 and CD8 single-positive (SP) thymocytes was blocked in mice lacking IKK1/2 in the T cell lineage. We found that IKK1/2-deficient thymocytes were specifically sensitized to TNF-induced cell death in vitro. Furthermore, the block in thymocyte development in IKK1/2-deficient mice could be rescued by blocking TNF with anti-TNF mAb or by ablation of TNFRI expression. These experiments reveal an essential role for TNF activation of NF-κB to promote the survival and development of single positive T cells in the thymus.
© 2016 Webb et al.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

