Directed evolution of a sphingomyelin flippase reveals mechanism of substrate backbone discrimination by a P4-ATPase
- PMID: 27432949
- PMCID: PMC4978280
- DOI: 10.1073/pnas.1525730113
Directed evolution of a sphingomyelin flippase reveals mechanism of substrate backbone discrimination by a P4-ATPase
Abstract
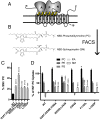
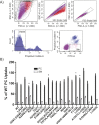
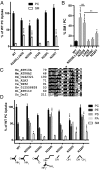
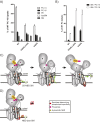
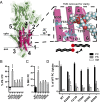
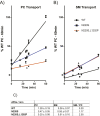
Phospholipid flippases in the type IV P-type ATPase (P4-ATPases) family establish membrane asymmetry and play critical roles in vesicular transport, cell polarity, signal transduction, and neurologic development. All characterized P4-ATPases flip glycerophospholipids across the bilayer to the cytosolic leaflet of the membrane, but how these enzymes distinguish glycerophospholipids from sphingolipids is not known. We used a directed evolution approach to examine the molecular mechanisms through which P4-ATPases discriminate substrate backbone. A mutagenesis screen in the yeast Saccharomyces cerevisiae has identified several gain-of-function mutations in the P4-ATPase Dnf1 that facilitate the transport of a novel lipid substrate, sphingomyelin. We found that a highly conserved asparagine (N220) in the first transmembrane segment is a key enforcer of glycerophospholipid selection, and specific substitutions at this site allow transport of sphingomyelin.
Keywords: P4-ATPase; directed evolution; membrane asymmetry; sphingomyelin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Vance DE, Vance JE. Biochemistry of Lipids, Lipoproteins and Membranes. 5th Ed Elsevier; Amsterdam: 2008.
-
- Devaux PF, Herrmann A. Transmembrane Dynamics of Lipids. Wiley; Hoboken, NJ: 2012.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

