Phylogeographic model selection leads to insight into the evolutionary history of four-eyed frogs
- PMID: 27432969
- PMCID: PMC4961127
- DOI: 10.1073/pnas.1601064113
Phylogeographic model selection leads to insight into the evolutionary history of four-eyed frogs
Abstract
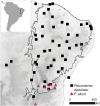
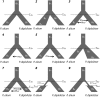
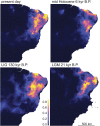
Phylogeographic research investigates biodiversity at the interface between populations and species, in a temporal and geographic context. Phylogeography has benefited from analytical approaches that allow empiricists to estimate parameters of interest from the genetic data (e.g., θ = 4Neμ, population divergence, gene flow), and the widespread availability of genomic data allow such parameters to be estimated with greater precision. However, the actual inferences made by phylogeographers remain dependent on qualitative interpretations derived from these parameters' values and as such may be subject to overinterpretation and confirmation bias. Here we argue in favor of using an objective approach to phylogeographic inference that proceeds by calculating the probability of multiple demographic models given the data and the subsequent ranking of these models using information theory. We illustrate this approach by investigating the diversification of two sister species of four-eyed frogs of northeastern Brazil using single nucleotide polymorphisms obtained via restriction-associated digest sequencing. We estimate the composite likelihood of the observed data given nine demographic models and then rank these models using Akaike information criterion. We demonstrate that estimating parameters under a model that is a poor fit to the data is likely to produce values that lead to spurious phylogeographic inferences. Our results strongly imply that identifying which parameters to estimate from a given system is a key step in the process of phylogeographic inference and is at least as important as being able to generate precise estimates of these parameters. They also illustrate that the incorporation of model uncertainty should be a component of phylogeographic hypothesis tests.
Keywords: Caatinga; Pleurodema; information theory; model selection; site frequency spectrum.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
An information-theoretical approach to phylogeography.Mol Ecol. 2009 Oct;18(20):4270-82. doi: 10.1111/j.1365-294X.2009.04327.x. Epub 2009 Sep 18. Mol Ecol. 2009. PMID: 19765225
-
Phylogeographic inference using Bayesian model comparison across a fragmented chorus frog species complex.Mol Ecol. 2015 Sep;24(18):4739-58. doi: 10.1111/mec.13343. Epub 2015 Sep 6. Mol Ecol. 2015. PMID: 26270246
-
Objective choice of phylogeographic models.Mol Phylogenet Evol. 2017 Nov;116:136-140. doi: 10.1016/j.ympev.2017.08.018. Epub 2017 Sep 5. Mol Phylogenet Evol. 2017. PMID: 28887148
-
Perspective: gene divergence, population divergence, and the variance in coalescence time in phylogeographic studies.Evolution. 2000 Dec;54(6):1839-54. doi: 10.1111/j.0014-3820.2000.tb01231.x. Evolution. 2000. PMID: 11209764 Review.
-
Assessing model adequacy leads to more robust phylogeographic inference.Trends Ecol Evol. 2022 May;37(5):402-410. doi: 10.1016/j.tree.2021.12.007. Epub 2022 Jan 10. Trends Ecol Evol. 2022. PMID: 35027224 Review.
Cited by
-
Phenotypes in phylogeography: Species' traits, environmental variation, and vertebrate diversification.Proc Natl Acad Sci U S A. 2016 Jul 19;113(29):8041-8. doi: 10.1073/pnas.1602237113. Proc Natl Acad Sci U S A. 2016. PMID: 27432983 Free PMC article. Review.
-
Ancient polymorphisms contribute to genome-wide variation by long-term balancing selection and divergent sorting in Boechera stricta.Genome Biol. 2019 Jun 21;20(1):126. doi: 10.1186/s13059-019-1729-9. Genome Biol. 2019. PMID: 31227026 Free PMC article.
-
A role of asynchrony of seasons in explaining genetic differentiation in a Neotropical toad.Heredity (Edinb). 2021 Oct;127(4):363-372. doi: 10.1038/s41437-021-00460-7. Epub 2021 Jul 24. Heredity (Edinb). 2021. PMID: 34304245 Free PMC article.
-
Using high-throughput sequencing to investigate the factors structuring genomic variation of a Mediterranean grasshopper of great conservation concern.Sci Rep. 2018 Sep 7;8(1):13436. doi: 10.1038/s41598-018-31775-x. Sci Rep. 2018. PMID: 30194365 Free PMC article.
-
In the light of evolution X: Comparative phylogeography.Proc Natl Acad Sci U S A. 2016 Jul 19;113(29):7957-61. doi: 10.1073/pnas.1604338113. Proc Natl Acad Sci U S A. 2016. PMID: 27432955 Free PMC article. No abstract available.
References
-
- Avise JC, et al. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Evol Syst. 1987;18:489–522.
-
- Knowles LL. The burgeoning field of statistical phylogeography. J Evol Biol. 2004;17(1):1–10. - PubMed
-
- Hickerson MJ, et al. Phylogeography’s past, present, and future: 10 years after Avise, 2000. Mol Phylogenet Evol. 2010;54(6):291–301. - PubMed
-
- Demesure B, Comps B, Petit RJ. Chloroplast DNA phylogeography of the common beech Fagus sylvatica L. in Europe. Evolution. 1996;50(6):2515–2520. - PubMed
-
- Bernatchez L, Wilson CC. Comparative phylogeography of Nearctic and Palearctic fishes. Mol Ecol. 1998;7(4):431–452.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

