Toward a paradigm shift in comparative phylogeography driven by trait-based hypotheses
- PMID: 27432974
- PMCID: PMC4961141
- DOI: 10.1073/pnas.1601069113
Toward a paradigm shift in comparative phylogeography driven by trait-based hypotheses
Abstract
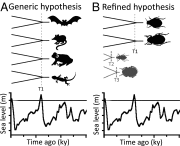
For three decades, comparative phylogeography has conceptually and methodologically relied on the concordance criterion for providing insights into the historical/biogeographic processes driving population genetic structure and divergence. Here we discuss how this emphasis, and the corresponding lack of methods for extracting information about biotic/intrinsic contributions to patterns of genetic variation, may bias our general understanding of the factors driving genetic structure. Specifically, this emphasis has promoted a tendency to attribute discordant phylogeographic patterns to the idiosyncracies of history, as well as an adherence to generic null expectations of concordance with reduced predictive power. We advocate that it is time for a paradigm shift in comparative phylogeography, especially given the limited utility of the concordance criterion as genomic data provide ever-increasing levels of resolution. Instead of adhering to the concordance-discordance dichotomy, comparative phylogeography needs to emphasize the contribution of taxon-specific traits that will determine whether concordance is a meaningful criterion for evaluating hypotheses or may predict discordant phylogeographic structure. Through reference to some case studies we illustrate how refined hypotheses based on taxon-specific traits can provide improved predictive frameworks to forecast species responses to climatic change or biogeographic barriers while gaining unique insights about the taxa themselves and their interactions with their environment. We outline a potential avenue toward a synthetic comparative phylogeographic paradigm that includes addressing some important conceptual and methodological challenges related to study design and application of model-based approaches for evaluating support of trait-based hypotheses under the proposed paradigm.
Keywords: Pleistocene; biogeography; coalescent modeling; ecological traits; statistical phylogeography.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Targeted Sampling and Target Capture: Assessing Phylogeographic Concordance with Genome-wide Data.Syst Biol. 2018 Nov 1;67(6):979-996. doi: 10.1093/sysbio/syy021. Syst Biol. 2018. PMID: 30339251
-
Phylogeography and biogeography of the lower Central American Neotropics: diversification between two continents and between two seas.Biol Rev Camb Philos Soc. 2014 Nov;89(4):767-90. doi: 10.1111/brv.12076. Epub 2014 Feb 3. Biol Rev Camb Philos Soc. 2014. PMID: 24495219 Review.
-
Integrating coalescent and ecological niche modeling in comparative phylogeography.Evolution. 2007 Jun;61(6):1439-54. doi: 10.1111/j.1558-5646.2007.00117.x. Evolution. 2007. PMID: 17542851
-
Species-specific responses to island connectivity cycles: refined models for testing phylogeographic concordance across a Mediterranean Pleistocene Aggregate Island Complex.Mol Ecol. 2015 Aug;24(16):4252-68. doi: 10.1111/mec.13305. Epub 2015 Jul 30. Mol Ecol. 2015. PMID: 26154606
-
Phenotypes in phylogeography: Species' traits, environmental variation, and vertebrate diversification.Proc Natl Acad Sci U S A. 2016 Jul 19;113(29):8041-8. doi: 10.1073/pnas.1602237113. Proc Natl Acad Sci U S A. 2016. PMID: 27432983 Free PMC article. Review.
Cited by
-
Size increase without genetic divergence in the Eurasian water shrew Neomys fodiens.Sci Rep. 2019 Nov 22;9(1):17375. doi: 10.1038/s41598-019-53891-y. Sci Rep. 2019. PMID: 31758027 Free PMC article.
-
Limited phylogeographic and genetic connectivity in Acacia species of low stature in an arid landscape.Ecol Evol. 2022 Jul 6;12(7):e9052. doi: 10.1002/ece3.9052. eCollection 2022 Jul. Ecol Evol. 2022. PMID: 35813908 Free PMC article.
-
East‒West genetic differentiation across the Indo-Burma hotspot: evidence from two closely related dioecious figs.BMC Plant Biol. 2023 Jun 16;23(1):321. doi: 10.1186/s12870-023-04324-6. BMC Plant Biol. 2023. PMID: 37322436 Free PMC article.
-
Evolutionary Genetics of Cacti: Research Biases, Advances and Prospects.Genes (Basel). 2022 Mar 1;13(3):452. doi: 10.3390/genes13030452. Genes (Basel). 2022. PMID: 35328006 Free PMC article. Review.
-
Full Bayesian Comparative Phylogeography from Genomic Data.Syst Biol. 2019 May 1;68(3):371-395. doi: 10.1093/sysbio/syy063. Syst Biol. 2019. PMID: 30239868 Free PMC article.
References
-
- Arbogast BS, Kenagy GJ. Comparative phylogeography as an integrative approach to historical biogeography. J Biogeogr. 2001;28(7):819–825.
-
- Zink RM. Methods in comparative phylogeography, and their application to studying evolution in the North American aridlands. Integr Comp Biol. 2002;42(5):953–959. - PubMed
-
- Avise JC. Phylogeography: The History and Formation of Species. Harvard Univ Press; Cambridge, MA: 2000.
-
- Avise JC. Molecular population structure and the biogeographic history of a regional fauna: A case history with lessons for conservation biology. Oikos. 1992;63(1):62–76.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

