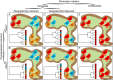
Phenotypes in phylogeography: Species' traits, environmental variation, and vertebrate diversification
- PMID: 27432983
- PMCID: PMC4961195
- DOI: 10.1073/pnas.1602237113
Phenotypes in phylogeography: Species' traits, environmental variation, and vertebrate diversification
Abstract
Almost 30 y ago, the field of intraspecific phylogeography laid the foundation for spatially explicit and genealogically informed studies of population divergence. With new methods and markers, the focus in phylogeography shifted to previously unrecognized geographic genetic variation, thus reducing the attention paid to phenotypic variation in those same diverging lineages. Although phenotypic differences among lineages once provided the main data for studies of evolutionary change, the mechanisms shaping phenotypic differentiation and their integration with intraspecific genetic structure have been underexplored in phylogeographic studies. However, phenotypes are targets of selection and play important roles in species performance, recognition, and diversification. Here, we focus on three questions. First, how can phenotypes elucidate mechanisms underlying concordant or idiosyncratic responses of vertebrate species evolving in shared landscapes? Second, what mechanisms underlie the concordance or discordance of phenotypic and phylogeographic differentiation? Third, how can phylogeography contribute to our understanding of functional phenotypic evolution? We demonstrate that the integration of phenotypic data extends the reach of phylogeography to explain the origin and maintenance of biodiversity. Finally, we stress the importance of natural history collections as sources of high-quality phenotypic data that span temporal and spatial axes.
Keywords: concordance; function; phenotype; phylogeography; trait.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Extensive phenotypic diversification coexists with little genetic divergence and a lack of population structure in the White Wagtail subspecies complex (Motacilla alba).J Evol Biol. 2018 Aug;31(8):1093-1108. doi: 10.1111/jeb.13305. Epub 2018 Jun 27. J Evol Biol. 2018. PMID: 29873425
-
Digging up the roots of an insular hotspot of genetic diversity: decoupled mito-nuclear histories in the evolution of the Corsican-Sardinian endemic lizard Podarcis tiliguerta.BMC Evol Biol. 2017 Mar 2;17(1):63. doi: 10.1186/s12862-017-0899-x. BMC Evol Biol. 2017. PMID: 28253846 Free PMC article.
-
Idiosyncratic responses to climate-driven forest fragmentation and marine incursions in reed frogs from Central Africa and the Gulf of Guinea Islands.Mol Ecol. 2017 Oct;26(19):5223-5244. doi: 10.1111/mec.14260. Epub 2017 Aug 24. Mol Ecol. 2017. PMID: 28753250
-
Comparative phylogeography of African savannah ungulates.Mol Ecol. 2012 Aug;21(15):3656-70. doi: 10.1111/j.1365-294X.2012.05650.x. Epub 2012 Jun 15. Mol Ecol. 2012. PMID: 22702960 Review.
-
Fitness of multidimensional phenotypes in dynamic adaptive landscapes.Trends Ecol Evol. 2015 Aug;30(8):487-96. doi: 10.1016/j.tree.2015.06.003. Epub 2015 Jun 26. Trends Ecol Evol. 2015. PMID: 26122484 Review.
Cited by
-
Evolutionary Genetics of Cacti: Research Biases, Advances and Prospects.Genes (Basel). 2022 Mar 1;13(3):452. doi: 10.3390/genes13030452. Genes (Basel). 2022. PMID: 35328006 Free PMC article. Review.
-
Annual aboveground carbon uptake enhancements from assisted gene flow in boreal black spruce forests are not long-lasting.Nat Commun. 2021 Feb 19;12(1):1169. doi: 10.1038/s41467-021-21222-3. Nat Commun. 2021. PMID: 33608515 Free PMC article.
-
Phylogeography and phenotypic wing shape variation in a damselfly across populations in Europe.BMC Ecol Evol. 2024 Feb 3;24(1):19. doi: 10.1186/s12862-024-02207-4. BMC Ecol Evol. 2024. PMID: 38308224 Free PMC article.
-
Hidden endemism, deep polyphyly, and repeated dispersal across the Isthmus of Tehuantepec: Diversification of the White-collared Seedeater complex (Thraupidae: Sporophila torqueola).Ecol Evol. 2018 Jan 12;8(3):1867-1881. doi: 10.1002/ece3.3799. eCollection 2018 Feb. Ecol Evol. 2018. PMID: 29435260 Free PMC article.
-
Inferring responses to climate dynamics from historical demography in neotropical forest lizards.Proc Natl Acad Sci U S A. 2016 Jul 19;113(29):7978-85. doi: 10.1073/pnas.1601063113. Proc Natl Acad Sci U S A. 2016. PMID: 27432951 Free PMC article.
References
-
- Avise JC, et al. Intraspecific phylogeography: The mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst. 1987;18:489–522.
-
- Buckley D. Toward an organismal, integrative, and iterative phylogeography. BioEssays. 2009;31(7):784–793. - PubMed
-
- Schneider CJ, Cunningham M, Moritz C. Comparative phylogeography and the history of endemic vertebrates in the Wet Tropics rainforests of Australia. Mol Ecol. 1998;7(4):487–498.
-
- Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405(6789):907–913. - PubMed
-
- Beheregaray LB. Twenty years of phylogeography: The state of the field and the challenges for the Southern Hemisphere. Mol Ecol. 2008;17(17):3754–3774. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources