The probability of monophyly of a sample of gene lineages on a species tree
- PMID: 27432988
- PMCID: PMC4961160
- DOI: 10.1073/pnas.1601074113
The probability of monophyly of a sample of gene lineages on a species tree
Abstract
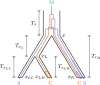
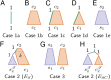
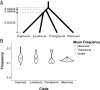
Monophyletic groups-groups that consist of all of the descendants of a most recent common ancestor-arise naturally as a consequence of descent processes that result in meaningful distinctions between organisms. Aspects of monophyly are therefore central to fields that examine and use genealogical descent. In particular, studies in conservation genetics, phylogeography, population genetics, species delimitation, and systematics can all make use of mathematical predictions under evolutionary models about features of monophyly. One important calculation, the probability that a set of gene lineages is monophyletic under a two-species neutral coalescent model, has been used in many studies. Here, we extend this calculation for a species tree model that contains arbitrarily many species. We study the effects of species tree topology and branch lengths on the monophyly probability. These analyses reveal new behavior, including the maintenance of nontrivial monophyly probabilities for gene lineage samples that span multiple species and even for lineages that do not derive from a monophyletic species group. We illustrate the mathematical results using an example application to data from maize and teosinte.
Keywords: coalescent theory; monophyly; phylogeography.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

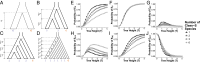
Similar articles
-
The Probability of Joint Monophyly of Samples of Gene Lineages for All Species in an Arbitrary Species Tree.J Comput Biol. 2022 Jul;29(7):679-703. doi: 10.1089/cmb.2021.0647. Epub 2022 May 11. J Comput Biol. 2022. PMID: 35544237 Free PMC article.
-
The probability of reciprocal monophyly of gene lineages in three and four species.Theor Popul Biol. 2019 Oct;129:133-147. doi: 10.1016/j.tpb.2018.04.004. Epub 2018 May 3. Theor Popul Biol. 2019. PMID: 29729946 Free PMC article.
-
The shapes of neutral gene genealogies in two species: probabilities of monophyly, paraphyly, and polyphyly in a coalescent model.Evolution. 2003 Jul;57(7):1465-77. doi: 10.1111/j.0014-3820.2003.tb00355.x. Evolution. 2003. PMID: 12940352
-
Phylogenetic Pattern, Evolutionary Processes and Species Delimitation in the Genus Echinococcus.Adv Parasitol. 2017;95:111-145. doi: 10.1016/bs.apar.2016.07.002. Epub 2016 Aug 13. Adv Parasitol. 2017. PMID: 28131362 Review.
-
Teosinte as a model system for population and ecological genomics.Trends Genet. 2012 Dec;28(12):606-15. doi: 10.1016/j.tig.2012.08.004. Epub 2012 Sep 27. Trends Genet. 2012. PMID: 23021022 Review.
Cited by
-
The R2R3-MYB gene family in banana (Musa acuminata): Genome-wide identification, classification and expression patterns.PLoS One. 2020 Oct 6;15(10):e0239275. doi: 10.1371/journal.pone.0239275. eCollection 2020. PLoS One. 2020. PMID: 33021974 Free PMC article.
-
Fungal species boundaries in the genomics era.Fungal Genet Biol. 2019 Oct;131:103249. doi: 10.1016/j.fgb.2019.103249. Epub 2019 Jul 4. Fungal Genet Biol. 2019. PMID: 31279976 Free PMC article. Review.
-
All galls are divided into three or more parts: recursive enumeration of labeled histories for galled trees.Algorithms Mol Biol. 2023 Feb 13;18(1):1. doi: 10.1186/s13015-023-00224-4. Algorithms Mol Biol. 2023. PMID: 36782318 Free PMC article.
-
Ages, sizes and (trees within) trees of taxa and of urns, from Yule to today.Philos Trans R Soc Lond B Biol Sci. 2025 Feb 13;380(1919):20230305. doi: 10.1098/rstb.2023.0305. Epub 2025 Feb 20. Philos Trans R Soc Lond B Biol Sci. 2025. PMID: 39976410
-
The Probability of Joint Monophyly of Samples of Gene Lineages for All Species in an Arbitrary Species Tree.J Comput Biol. 2022 Jul;29(7):679-703. doi: 10.1089/cmb.2021.0647. Epub 2022 May 11. J Comput Biol. 2022. PMID: 35544237 Free PMC article.
References
-
- Neigel J, Avise J. Phylogenetic relationships of mitochondrial DNA under various demographic models of speciation. In: Karlin S, Nevo E, editors. Evolutionary Processes and Theory. Academic Press; San Diego: 1986. pp. 515–534.
-
- Rosenberg NA. The shapes of neutral gene genealogies in two species: Probabilities of monophyly, paraphyly, and polyphyly in a coalescent model. Evolution. 2003;57(7):1465–1477. - PubMed
-
- Degnan JH, Salter LA. Gene tree distributions under the coalescent process. Evolution. 2005;59(1):24–37. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

