Plastidial metabolite MEcPP induces a transcriptionally centered stress-response hub via the transcription factor CAMTA3
- PMID: 27432993
- PMCID: PMC4978302
- DOI: 10.1073/pnas.1602582113
Plastidial metabolite MEcPP induces a transcriptionally centered stress-response hub via the transcription factor CAMTA3
Abstract
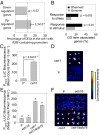
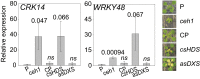
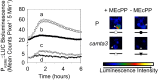
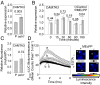
The general stress response (GSR) is an evolutionarily conserved rapid and transient transcriptional reprograming of genes central for transducing environmental signals into cellular responses, leading to metabolic and physiological readjustments to cope with prevailing conditions. Defining the regulatory components of the GSR will provide crucial insight into the design principles of early stress-response modules and their role in orchestrating master regulators of adaptive responses. Overaccumulation of methylerythritol cyclodiphosphate (MEcPP), a bifunctional chemical entity serving as both a precursor of isoprenoids produced by the plastidial methylerythritol phosphate (MEP) pathway and a stress-specific retrograde signal, in ceh1 (constitutively expressing hydroperoxide lyase1)-mutant plants leads to large-scale transcriptional alterations. Bioinformatic analyses of microarray data in ceh1 plants established the overrepresentation of a stress-responsive cis element and key GSR marker, the rapid stress response element (RSRE), in the promoters of robustly induced genes. ceh1 plants carrying an established 4×RSRE:Luciferase reporter for monitoring the GSR support constitutive activation of the response in this mutant background. Genetics and pharmacological approaches confirmed the specificity of MEcPP in RSRE induction via the transcription factor CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 3 (CAMTA3), in a calcium-dependent manner. Moreover, CAMTA3-dependent activation of IRE1a (inositol-requiring protein-1) and bZIP60 (basic leucine zipper 60), two RSRE containing unfolded protein-response genes, bridges MEcPP-mediated GSR induction to the potentiation of protein-folding homeostasis in the endoplasmic reticulum. These findings introduce the notion of transcriptional regulation by a key plastidial retrograde signaling metabolite that induces nuclear GSR, thereby offering a window into the role of interorgannellar communication in shaping cellular adaptive responses.
Keywords: CAMTA3; GSR; MEcPP; RSRE; retrograde signals.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Kültz D. Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol. 2005;67:225–257. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

