Long Non-coding RNA BGas Regulates the Cystic Fibrosis Transmembrane Conductance Regulator
- PMID: 27434588
- PMCID: PMC5023374
- DOI: 10.1038/mt.2016.112
Long Non-coding RNA BGas Regulates the Cystic Fibrosis Transmembrane Conductance Regulator
Abstract
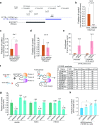
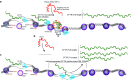
Cystic fibrosis (CF) is a life-shortening genetic disease. The root cause of CF is heritable recessive mutations that affect the cystic fibrosis transmembrance conductance regulator (CFTR) gene and the subsequent expression and activity of encoded ion channels at the cell surface. We show that CFTR is regulated transcriptionally by the actions of a novel long noncoding RNA (lncRNA), designated as BGas, that emanates from intron 11 of the CFTR gene and is expressed in the antisense orientation relative to the protein coding sense strand. We find that BGas functions in concert with several proteins including HMGA1, HMGB1, and WIBG to modulate the local chromatin and DNA architecture of intron 11 of the CFTR gene and thereby affects transcription. Suppression of BGas or its associated proteins results in a gain of both CFTR expression and chloride ion function. The observations described here highlight a previously underappreciated mechanism of transcriptional control and suggest that BGas may serve as a therapeutic target for specifically activating expression of CFTR.
Figures

References
-
- Kerem, B, Rommens, JM, Buchanan, JA, Markiewicz, D, Cox, TK, Chakravarti, A et al. (1989). Identification of the cystic fibrosis gene: genetic analysis. Science 245: 1073–1080. - PubMed
-
- Riordan, JR, Rommens, JM, Kerem, B, Alon, N, Rozmahel, R, Grzelczak, Z et al. (1989). Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245: 1066–1073. - PubMed
-
- Rommens, JM, Iannuzzi, MC, Kerem, B, Drumm, ML, Melmer, G, Dean, M et al. (1989). Identification of the cystic fibrosis gene: chromosome walking and jumping. Science 245: 1059–1065. - PubMed
-
- Rowe, SM, Miller, S and Sorscher, EJ (2005). Cystic fibrosis. N Engl J Med 352: 1992–2001. - PubMed
-
- Bombieri, C, Seia, M and Castellani, C (2015). Genotypes and phenotypes in cystic fibrosis and cystic fibrosis transmembrane regulator-related disorders. Semin Respir Crit Care Med 36: 180–193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

