Multiple capsid-stabilizing interactions revealed in a high-resolution structure of an emerging picornavirus causing neonatal sepsis
- PMID: 27435188
- PMCID: PMC4961769
- DOI: 10.1038/ncomms11387
Multiple capsid-stabilizing interactions revealed in a high-resolution structure of an emerging picornavirus causing neonatal sepsis
Abstract
The poorly studied picornavirus, human parechovirus 3 (HPeV3) causes neonatal sepsis with no therapies available. Our 4.3-Å resolution structure of HPeV3 on its own and at 15 Å resolution in complex with human monoclonal antibody Fabs demonstrates the expected picornavirus capsid structure with three distinct features. First, 25% of the HPeV3 RNA genome in 60 sites is highly ordered as confirmed by asymmetric reconstruction, and interacts with conserved regions of the capsid proteins VP1 and VP3. Second, the VP0 N terminus stabilizes the capsid inner surface, in contrast to other picornaviruses where on expulsion as VP4, it forms an RNA translocation channel. Last, VP1's hydrophobic pocket, the binding site for the antipicornaviral drug, pleconaril, is blocked and thus inappropriate for antiviral development. Together, these results suggest a direction for development of neutralizing antibodies, antiviral drugs based on targeting the RNA-protein interactions and dissection of virus assembly on the basis of RNA nucleation.
Conflict of interest statement
AIMM Therapeutics employed T.B. and A.Q.B. The remaining authors declare no competing financial interests.
Figures

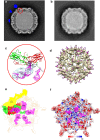
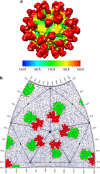
Similar articles
-
The Structure of Human Parechovirus 1 Reveals an Association of the RNA Genome with the Capsid.J Virol. 2015 Nov 18;90(3):1377-86. doi: 10.1128/JVI.02346-15. Print 2016 Feb 1. J Virol. 2015. PMID: 26581987 Free PMC article.
-
A 2.8-Angstrom-Resolution Cryo-Electron Microscopy Structure of Human Parechovirus 3 in Complex with Fab from a Neutralizing Antibody.J Virol. 2019 Feb 5;93(4):e01597-18. doi: 10.1128/JVI.01597-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30463974 Free PMC article.
-
Intrinsically-disordered N-termini in human parechovirus 1 capsid proteins bind encapsidated RNA.Sci Rep. 2018 Apr 11;8(1):5820. doi: 10.1038/s41598-018-23552-7. Sci Rep. 2018. PMID: 29643409 Free PMC article.
-
Evolutionary and Structural Overview of Human Picornavirus Capsid Antibody Evasion.Front Cell Infect Microbiol. 2019 Aug 20;9:283. doi: 10.3389/fcimb.2019.00283. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31482072 Free PMC article. Review.
-
Hepatitis A Virus Capsid Structure.Cold Spring Harb Perspect Med. 2019 May 1;9(5):a031807. doi: 10.1101/cshperspect.a031807. Cold Spring Harb Perspect Med. 2019. PMID: 30037986 Free PMC article. Review.
Cited by
-
CryoVirusDB: A Labeled Cryo-EM Image Dataset for AI-Driven Virus Particle Picking.bioRxiv [Preprint]. 2023 Dec 26:2023.12.25.573312. doi: 10.1101/2023.12.25.573312. bioRxiv. 2023. PMID: 38234823 Free PMC article. Preprint.
-
The In Silico Prediction of Hotspot Residues that Contribute to the Structural Stability of Subunit Interfaces of a Picornavirus Capsid.Viruses. 2020 Mar 31;12(4):387. doi: 10.3390/v12040387. Viruses. 2020. PMID: 32244486 Free PMC article.
-
Stability of SARS-CoV-2 and other airborne viruses under different stress conditions.Arch Virol. 2022 Jan;167(1):183-187. doi: 10.1007/s00705-021-05293-7. Epub 2021 Nov 2. Arch Virol. 2022. PMID: 34727217 Free PMC article.
-
Swin-cryoEM: Multi-class cryo-electron micrographs single particle mixed detection method.PLoS One. 2024 Apr 9;19(4):e0298287. doi: 10.1371/journal.pone.0298287. eCollection 2024. PLoS One. 2024. PMID: 38593135 Free PMC article.
-
Strain-dependent neutralization reveals antigenic variation of human parechovirus 3.Sci Rep. 2017 Sep 21;7(1):12075. doi: 10.1038/s41598-017-12458-5. Sci Rep. 2017. PMID: 28935894 Free PMC article.
References
-
- Wolthers K. C. et al.. Human parechoviruses as an important viral cause of sepsislike illness and meningitis in young children. Clin. Infect. Dis. 47, 358–363 (2008). - PubMed
-
- Wildenbeest J. G., Harvala H., Pajkrt D. & Wolthers K. C. The need for treatment against human parechoviruses: how, why and when? Expert Rev. Anti Infect. Ther. 8, 1417–1429 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

