The unfolded protein response genes in human osteoarthritic chondrocytes: PERK emerges as a potential therapeutic target
- PMID: 27435272
- PMCID: PMC4952234
- DOI: 10.1186/s13075-016-1070-6
The unfolded protein response genes in human osteoarthritic chondrocytes: PERK emerges as a potential therapeutic target
Abstract
Background: The unfolded protein response (UPR) is activated following an endoplasmic reticulum (ER) stress. The aim of this study was to investigate the global expression of UPR genes in human OA chondrocytes in induced (I)-UPR conditions, and to explore the regulation and role of the UPR genes in homeostatic (H)-UPR conditions in human normal and OA chondrocytes.
Methods: Gene expression was determined by PCR array and qPCR. Protein production in cartilage was determined by immunohistochemistry, gene silencing by specific siRNAs, and gene regulation by treating chondrocytes with cytokines and growth factors associated with cartilage pathobiology.
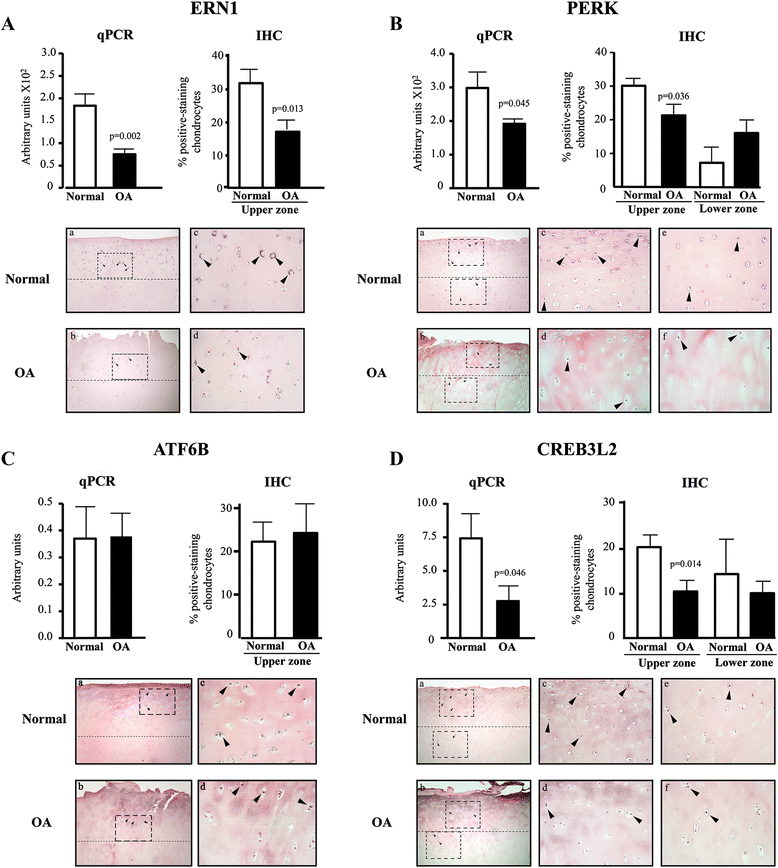
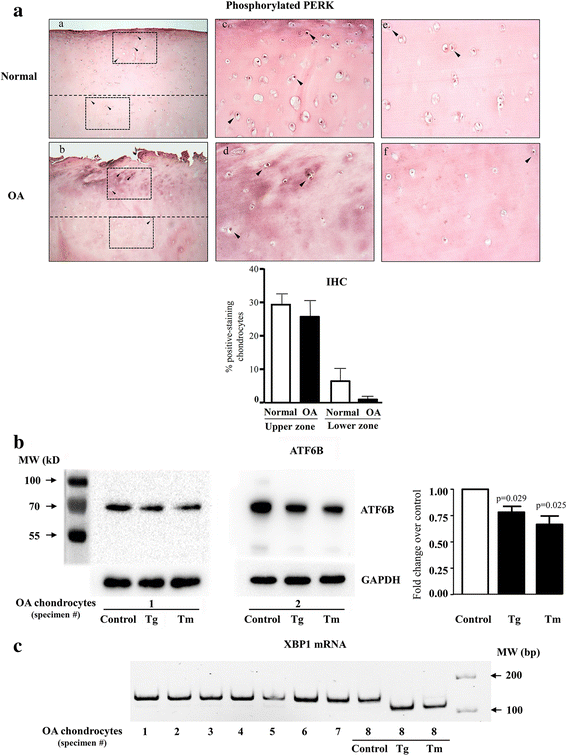
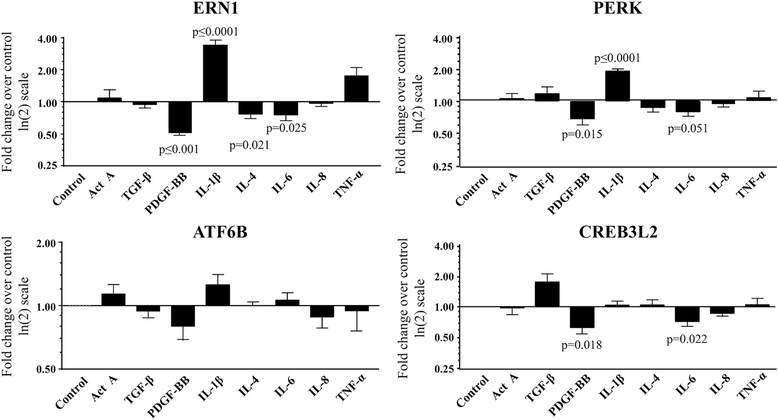
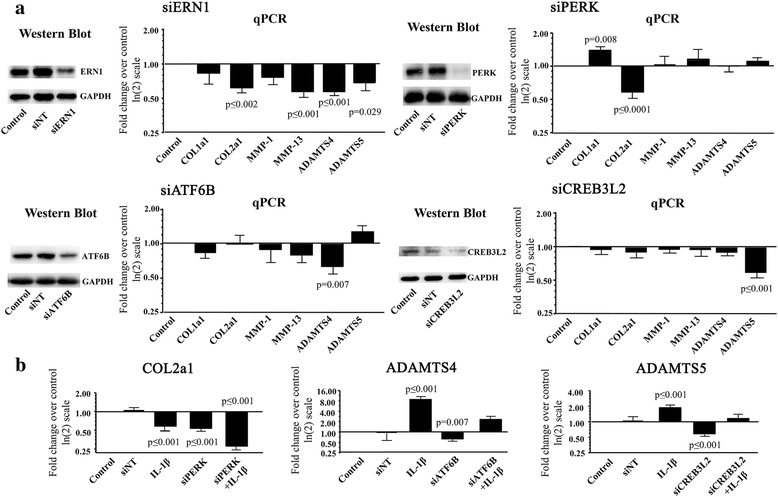
Results: Several UPR genes, among them ERN1, PERK, and CREB3L2 were downregulated in OA compared to normal chondrocytes at both the mRNA and protein levels, but the ER stress response triggered by thapsigargin or tunicamycin treatment was similar in normal and OA chondrocytes. The activation of ER stress sensors (phosphorylated PERK, cleavage of ATF6B, and the spliced mRNA forms of XBP1) was not significantly increased in OA chondrocytes/cartilage. PDGF-BB and IL-6 significantly downregulated the expression of ERN1, PERK, and CREB3L2, but not that of ATF6B. Silencing experiments done under conditions of no ER stress (physiological conditions) revealed that decreasing ERN1 expression led to decreased COL2a1, MMP-13, ADAMTS4 and ADAMTS5 expression, while decreasing CREB3L2 and ATF6B led to decreased ADAMTS5 and ADAMTS4 expression, respectively. Importantly, the downregulation of PERK expression increased COL1a1 and suppressed COL2a1 expression.
Conclusions: Although the level of ER stress is not significantly increased in OA chondrocytes, these cells respond strongly to an acute ER stress despite the decreased expression of ERN1, PERK, and CREB3L2. Emerging findings revealed for the first time that these genes play a role in cartilage biology in conditions where an acute ER stress response is not triggered and OA is not characterized by an overall basal activation of the ER stress response. Importantly, these findings identify PERK as a potential target for new OA treatment avenues.
Keywords: Chondrocyte; ER stress; Osteoarthritis; PERK; Unfolded protein response.
Figures
References
-
- Reiling JH, Clish CB, Carette JE, Varadarajan M, Brummelkamp TR, Sabatini DM. A haploid genetic screen identifies the major facilitator domain containing 2A (MFSD2A) transporter as a key mediator in the response to tunicamycin. Proc Natl Acad Sci U S A. 2011;108(29):11756–65. doi: 10.1073/pnas.1018098108. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

