Exome sequencing in mostly consanguineous Arab families with neurologic disease provides a high potential molecular diagnosis rate
- PMID: 27435318
- PMCID: PMC4950750
- DOI: 10.1186/s12920-016-0208-3
Exome sequencing in mostly consanguineous Arab families with neurologic disease provides a high potential molecular diagnosis rate
Abstract
Background: Neurodevelopment is orchestrated by a wide range of genes, and the genetic causes of neurodevelopmental disorders are thus heterogeneous. We applied whole exome sequencing (WES) for molecular diagnosis and in silico analysis to identify novel disease gene candidates in a cohort from Saudi Arabia with primarily Mendelian neurologic diseases.
Methods: We performed WES in 31 mostly consanguineous Arab families and analyzed both single nucleotide and copy number variants (CNVs) from WES data. Interaction/expression network and pathway analyses, as well as paralog studies were utilized to investigate potential pathogenicity and disease association of novel candidate genes. Additional cases for candidate genes were identified through the clinical WES database at Baylor Miraca Genetics Laboratories and GeneMatcher.
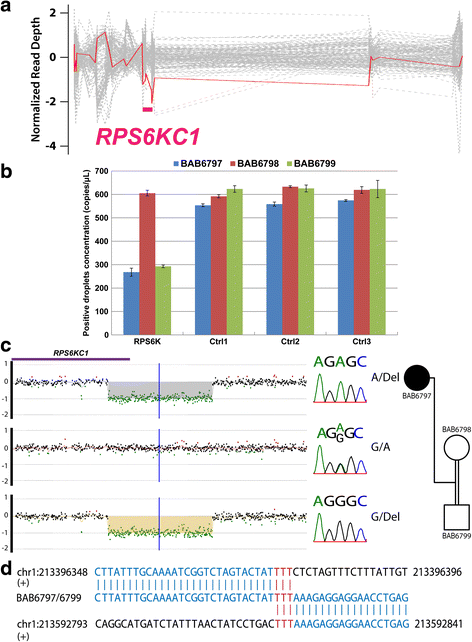
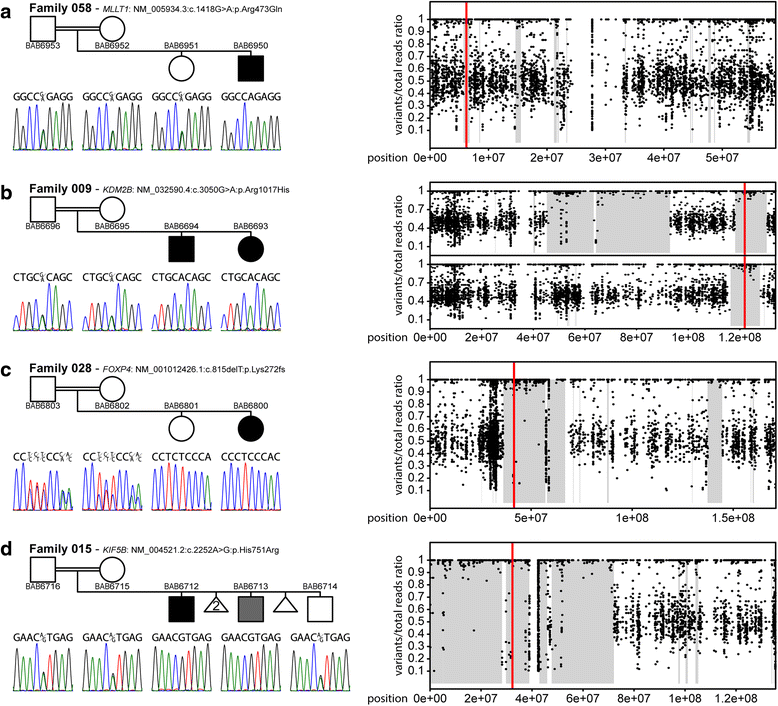
Results: We found known pathogenic or novel variants in known disease genes with phenotypic expansion in 6 families, disease-associated CNVs in 2 families, and 12 novel disease gene candidates in 11 families, including KIF5B, GRM7, FOXP4, MLLT1, and KDM2B. Overall, a potential molecular diagnosis was provided by variants in known disease genes in 17 families (54.8 %) and by novel candidate disease genes in an additional 11 families, making the potential molecular diagnostic rate ~90 %.
Conclusions: Molecular diagnostic rate from WES is improved by exome-predicted CNVs. Novel candidate disease gene discovery is facilitated by paralog studies and through the use of informatics tools and available databases to identify additional evidence for pathogenicity.
Trial registration: Not applicable.
Keywords: Copy Number Variants (CNV); Developmental Delay/Intellectual Disability (DD/ID); GRM7; Neurodevelopment; Whole exome sequencing (WES).
Figures


References
-
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, Abajian C, Beckmann CF, Bernard A, Bertagnolli D, Boe AF, Cartagena PM, Chakravarty MM, Chapin M, Chong J, Dalley RA, Daly BD, Dang C, Datta S, Dee N, Dolbeare TA, Faber V, Feng D, Fowler DR, Goldy J, Gregor BW, Haradon Z, Haynor DR, Hohmann JG, Horvath S, Howard RE, Jeromin A, Jochim JM, Kinnunen M, Lau C, Lazarz ET, Lee C, Lemon TA, Li L, Li Y, Morris JA, Overly CC, Parker PD, Parry SE, Reding M, Royall JJ, Schulkin J, Sequeira PA, Slaughterbeck CR, Smith SC, Sodt AJ, Sunkin SM, Swanson BE, Vawter MP, Williams D, Wohnoutka P, Zielke HR, Geschwind DH, Hof PR, Smith SM, Koch C, Grant SG, Jones AR. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489(7416):391–9. doi: 10.1038/nature11405. - DOI - PMC - PubMed
-
- Drachman DA. Do we have brain to spare? Neurology. 2005;64(12):2004–5. doi: 10.1212/01.WNL.0000166914.38327.BB. - DOI - PubMed
-
- Alazami AM, Patel N, Shamseldin HE, Anazi S, Al-Dosari MS, Alzahrani F, Hijazi H, Alshammari M, Aldahmesh MA, Salih MA, Faqeih E, Alhashem A, Bashiri FA, Al-Owain M, Kentab AY, Sogaty S, Al Tala S, Temsah MH, Tulbah M, Aljelaify RF, Alshahwan SA, Seidahmed MZ, Alhadid AA, Aldhalaan H, AlQallaf F, Kurdi W, Alfadhel M, Babay Z, Alsogheer M, Kaya N, Al-Hassnan ZN, Abdel-Salam GM, Al-Sannaa N, Al Mutairi F, El Khashab HY, Bohlega S, Jia X, Nguyen HC, Hammami R, Adly N, Mohamed JY, Abdulwahab F, Ibrahim N, Naim EA, Al-Younes B, Meyer BF, Hashem M, Shaheen R, Xiong Y, Abouelhoda M, Aldeeri AA, Monies DM, Alkuraya FS. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 2015;10(2):148–61. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

