Archaeal Haloarcula californiae Icosahedral Virus 1 Highlights Conserved Elements in Icosahedral Membrane-Containing DNA Viruses from Extreme Environments
- PMID: 27435460
- PMCID: PMC4958249
- DOI: 10.1128/mBio.00699-16
Archaeal Haloarcula californiae Icosahedral Virus 1 Highlights Conserved Elements in Icosahedral Membrane-Containing DNA Viruses from Extreme Environments
Abstract
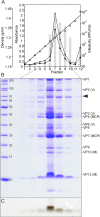
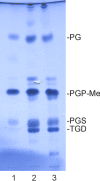
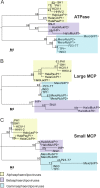
Despite their high genomic diversity, all known viruses are structurally constrained to a limited number of virion morphotypes. One morphotype of viruses infecting bacteria, archaea, and eukaryotes is the tailless icosahedral morphotype with an internal membrane. Although it is considered an abundant morphotype in extreme environments, only seven such archaeal viruses are known. Here, we introduce Haloarcula californiae icosahedral virus 1 (HCIV-1), a halophilic euryarchaeal virus originating from salt crystals. HCIV-1 also retains its infectivity under low-salinity conditions, showing that it is able to adapt to environmental changes. The release of progeny virions resulting from cell lysis was evidenced by reduced cellular oxygen consumption, leakage of intracellular ATP, and binding of an indicator ion to ruptured cell membranes. The virion contains at least 12 different protein species, lipids selectively acquired from the host cell membrane, and a 31,314-bp-long linear double-stranded DNA (dsDNA). The overall genome organization and sequence show high similarity to the genomes of archaeal viruses in the Sphaerolipoviridae family. Phylogenetic analysis based on the major conserved components needed for virion assembly-the major capsid proteins and the packaging ATPase-placed HCIV-1 along with the alphasphaerolipoviruses in a distinct, well-supported clade. On the basis of its virion morphology and sequence similarities, most notably, those of its core virion components, we propose that HCIV-1 is a member of the PRD1-adenovirus structure-based lineage together with other sphaerolipoviruses. This addition to the lineage reinforces the notion of the ancient evolutionary links observed between the viruses and further highlights the limits of the choices found in nature for formation of a virion.
Importance: Under conditions of extreme salinity, the majority of the organisms present are archaea, which encounter substantial selective pressure, being constantly attacked by viruses. Regardless of the enormous viral sequence diversity, all known viruses can be clustered into a few structure-based viral lineages based on their core virion components. Our description of a new halophilic virus-host system adds significant insights into the largely unstudied field of archaeal viruses and, in general, of life under extreme conditions. Comprehensive molecular characterization of HCIV-1 shows that this icosahedral internal membrane-containing virus exhibits conserved elements responsible for virion organization. This places the virus neatly in the PRD1-adenovirus structure-based lineage. HCIV-1 further highlights the limited diversity of virus morphotypes despite the astronomical number of viruses in the biosphere. The observed high conservation in the core virion elements should be considered in addressing such fundamental issues as the origin and evolution of viruses and their interplay with their hosts.
Copyright © 2016 Demina et al.
Figures



References
-
- Maaty WS, Ortmann AC, Dlakić M, Schulstad K, Hilmer JK, Liepold L, Weidenheft B, Khayat R, Douglas T, Young MJ, Bothner B. 2006. Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life. J Virol 80:7625–7635. doi: 10.1128/JVI.00522-06. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
