Prospects of microbial cell factories developed through systems metabolic engineering
- PMID: 27435545
- PMCID: PMC4993179
- DOI: 10.1111/1751-7915.12385
Prospects of microbial cell factories developed through systems metabolic engineering
Abstract
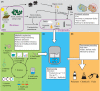
While academic-level studies on metabolic engineering of microorganisms for production of chemicals and fuels are ever growing, a significantly lower number of such production processes have reached commercial-scale. In this work, we review the challenges associated with moving from laboratory-scale demonstration of microbial chemical or fuel production to actual commercialization, focusing on key requirements on the production organism that need to be considered during the metabolic engineering process. Metabolic engineering strategies should take into account techno-economic factors such as the choice of feedstock, the product yield, productivity and titre, and the cost effectiveness of midstream and downstream processes. Also, it is important to develop an industrial strain through metabolic engineering for pathway construction and flux optimization together with increasing tolerance to products and inhibitors present in the feedstock, and ensuring genetic stability and strain robustness under actual fermentation conditions.
© 2016 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd and Society for Applied Microbiology.
Figures
References
-
- Alper, H. , Moxley, J. , Nevoigt, E. , Fink, G.R. , and Stephanopoulos, G.N. (2006) Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314: 1565–1568. - PubMed
-
- Bonde, M.T. , Pedersen, M. , Klausen, M.S. , Jensen, S.I. , Wulff, T. , Harrison, S. , et al (2016) Predictable tuning of protein expression in bacteria. Nat Methods 13: 233–236. - PubMed
-
- Ceroni, F. , Algar, R. , Stan, G.‐B. , and Ellis, T. (2015) Quantifying cellular capacity identifies gene expression designs with reduced burden. Nat Methods 12: 415–418. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

