Identification of Oligosaccharides in Feces of Breast-fed Infants and Their Correlation with the Gut Microbial Community
- PMID: 27435585
- PMCID: PMC5013312
- DOI: 10.1074/mcp.M116.060665
Identification of Oligosaccharides in Feces of Breast-fed Infants and Their Correlation with the Gut Microbial Community
Abstract
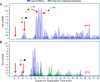
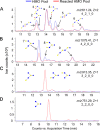
Glycans in breast milk are abundant and found as either free oligosaccharides or conjugated to proteins and lipids. Free human milk oligosaccharides (HMOs) function as prebiotics by stimulating the growth of beneficial bacteria while preventing the binding of harmful bacteria to intestinal epithelial cells. Bacteria have adapted to the glycan-rich environment of the gut by developing enzymes that catabolize glycans. The decrease in HMOs and the increase in glycan digestion products give indications of the active enzymes in the microbial population. In this study, we quantitated the disappearance of intact HMOs and characterized the glycan digestion products in the gut that are produced by the action of microbial enzymes on HMOs and glycoconjugates from breast milk. Oligosaccharides from fecal samples of exclusively breast-fed infants were extracted and profiled using nanoLC-MS. Intact HMOs were found in the fecal samples, additionally, other oligosaccharides were found corresponding to degraded HMOs and non-HMO based compounds. The latter compounds were fragments of N-glycans released through the cleavage of the linkage to the asparagine residue and through cleavage of the chitobiose core of the N-glycan. Marker gene sequencing of the fecal samples revealed bifidobacteria as the dominant inhabitants of the infant gastrointestinal tracts. A glycosidase from Bifidobacterium longum subsp. longum was then expressed to digest HMOs in vitro, which showed that the digested oligosaccharides in feces corresponded to the action of glycosidases on HMOs. Similar expression of endoglycosidases also showed that N-glycans were released by bacterial enzymes. Although bifidobacteria may dominate the gut, it is possible that specific minority species are also responsible for the major products observed in feces. Nonetheless, the enzymatic activity correlated well with the known glycosidases in the respective bacteria, suggesting a direct relationship between microbial abundances and catabolic activity.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




Similar articles
-
Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: a proof-of-concept study.J Proteome Res. 2015 Jan 2;14(1):491-502. doi: 10.1021/pr500759e. Epub 2014 Oct 28. J Proteome Res. 2015. PMID: 25300177 Free PMC article.
-
Varied Pathways of Infant Gut-Associated Bifidobacterium to Assimilate Human Milk Oligosaccharides: Prevalence of the Gene Set and Its Correlation with Bifidobacteria-Rich Microbiota Formation.Nutrients. 2019 Dec 26;12(1):71. doi: 10.3390/nu12010071. Nutrients. 2019. PMID: 31888048 Free PMC article. Review.
-
Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum.Sci Rep. 2018 Sep 18;8(1):13958. doi: 10.1038/s41598-018-32080-3. Sci Rep. 2018. PMID: 30228375 Free PMC article.
-
Linking Human Milk Oligosaccharides, Infant Fecal Community Types, and Later Risk To Require Antibiotics.mBio. 2020 Mar 17;11(2):e03196-19. doi: 10.1128/mBio.03196-19. mBio. 2020. PMID: 32184252 Free PMC article. Clinical Trial.
-
Linking human milk oligosaccharide metabolism and early life gut microbiota: bifidobacteria and beyond.Microbiol Mol Biol Rev. 2024 Mar 27;88(1):e0009423. doi: 10.1128/mmbr.00094-23. Epub 2024 Jan 11. Microbiol Mol Biol Rev. 2024. PMID: 38206006 Free PMC article. Review.
Cited by
-
Bifidobacterium longum and microbiome maturation modify a nutrient intervention for stunting in Zimbabwean infants.EBioMedicine. 2024 Oct;108:105362. doi: 10.1016/j.ebiom.2024.105362. Epub 2024 Sep 27. EBioMedicine. 2024. PMID: 39341154 Free PMC article.
-
The Infant Gut Commensal Bacteroides dorei Presents a Generalized Transcriptional Response to Various Human Milk Oligosaccharides.Front Cell Infect Microbiol. 2022 Mar 18;12:854122. doi: 10.3389/fcimb.2022.854122. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35372092 Free PMC article.
-
Alleviation of Intestinal Inflammation by Oral Supplementation With 2-Fucosyllactose in Mice.Front Microbiol. 2019 Jun 19;10:1385. doi: 10.3389/fmicb.2019.01385. eCollection 2019. Front Microbiol. 2019. PMID: 31275292 Free PMC article.
-
Persistence of Supplemented Bifidobacterium longum subsp. infantis EVC001 in Breastfed Infants.mSphere. 2017 Dec 6;2(6):e00501-17. doi: 10.1128/mSphere.00501-17. eCollection 2017 Nov-Dec. mSphere. 2017. PMID: 29242832 Free PMC article.
-
In Love with Shaping You-Influential Factors on the Breast Milk Content of Human Milk Oligosaccharides and Their Decisive Roles for Neonatal Development.Nutrients. 2020 Nov 20;12(11):3568. doi: 10.3390/nu12113568. Nutrients. 2020. PMID: 33233832 Free PMC article. Review.
References
-
- Ruhaak L. R., and Lebrilla C. B. (2012) Analysis and role of oligosaccharides in milk. BMB Reports 45, 442–451 - PubMed
-
- Liao Y., Alvarado R., Phinney B., and Lonnerdal B. (2011) Proteomic characterization of human milk whey proteins during a twelve-month lactation period. J. Proteome Res. 10, 1746–1754 - PubMed
-
- Kunz C., and Rudloff S. (1993) Biological functions of oligosaccharides in human milk. Acta Paediatr. 82, 903–912 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

