Monochromatic multicomponent fluorescence sedimentation velocity for the study of high-affinity protein interactions
- PMID: 27436096
- PMCID: PMC4985284
- DOI: 10.7554/eLife.17812
Monochromatic multicomponent fluorescence sedimentation velocity for the study of high-affinity protein interactions
Abstract
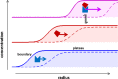
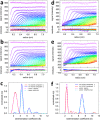
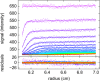
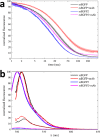
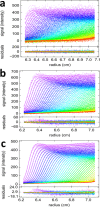
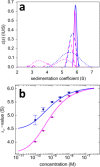
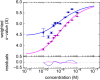
The dynamic assembly of multi-protein complexes underlies fundamental processes in cell biology. A mechanistic understanding of assemblies requires accurate measurement of their stoichiometry, affinity and cooperativity, and frequently consideration of multiple co-existing complexes. Sedimentation velocity analytical ultracentrifugation equipped with fluorescence detection (FDS-SV) allows the characterization of protein complexes free in solution with high size resolution, at concentrations in the nanomolar and picomolar range. Here, we extend the capabilities of FDS-SV with a single excitation wavelength from single-component to multi-component detection using photoswitchable fluorescent proteins (psFPs). We exploit their characteristic quantum yield of photo-switching to imprint spatio-temporal modulations onto the sedimentation signal that reveal different psFP-tagged protein components in the mixture. This novel approach facilitates studies of heterogeneous multi-protein complexes at orders of magnitude lower concentrations and for higher-affinity systems than previously possible. Using this technique we studied high-affinity interactions between the amino-terminal domains of GluA2 and GluA3 AMPA receptors.
Keywords: analytical ultracentrifugation; biophysics; hydrodynamics; photoswitchable fluorescent proteins; protein interactions; structural biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures


Similar articles
-
Analysis of protein interactions with picomolar binding affinity by fluorescence-detected sedimentation velocity.Anal Chem. 2014 Mar 18;86(6):3181-7. doi: 10.1021/ac500093m. Epub 2014 Mar 5. Anal Chem. 2014. PMID: 24552356 Free PMC article.
-
Analysis of high affinity self-association by fluorescence optical sedimentation velocity analytical ultracentrifugation of labeled proteins: opportunities and limitations.PLoS One. 2013 Dec 17;8(12):e83439. doi: 10.1371/journal.pone.0083439. eCollection 2013. PLoS One. 2013. PMID: 24358283 Free PMC article.
-
Studying multiprotein complexes by multisignal sedimentation velocity analytical ultracentrifugation.Proc Natl Acad Sci U S A. 2005 Jan 4;102(1):81-6. doi: 10.1073/pnas.0408399102. Epub 2004 Dec 21. Proc Natl Acad Sci U S A. 2005. PMID: 15613487 Free PMC article.
-
Modern analytical ultracentrifugation in protein science: a tutorial review.Protein Sci. 2002 Sep;11(9):2067-79. doi: 10.1110/ps.0207702. Protein Sci. 2002. PMID: 12192063 Free PMC article. Review.
-
Analytical ultracentrifugation for the study of protein association and assembly.Curr Opin Chem Biol. 2006 Oct;10(5):430-6. doi: 10.1016/j.cbpa.2006.08.017. Epub 2006 Aug 28. Curr Opin Chem Biol. 2006. PMID: 16935549 Review.
Cited by
-
Sedimentation of Reversibly Interacting Macromolecules with Changes in Fluorescence Quantum Yield.Biophys J. 2017 Apr 11;112(7):1374-1382. doi: 10.1016/j.bpj.2017.02.020. Biophys J. 2017. PMID: 28402880 Free PMC article.
-
Nucleic acid-induced dimerization of HIV-1 Gag protein.J Biol Chem. 2019 Nov 8;294(45):16480-16493. doi: 10.1074/jbc.RA119.010580. Epub 2019 Sep 30. J Biol Chem. 2019. PMID: 31570521 Free PMC article.
-
Measuring macromolecular size distributions and interactions at high concentrations by sedimentation velocity.Nat Commun. 2018 Oct 24;9(1):4415. doi: 10.1038/s41467-018-06902-x. Nat Commun. 2018. PMID: 30356043 Free PMC article.
-
Sedimentation velocity analytical ultracentrifugation for characterization of therapeutic antibodies.Biophys Rev. 2018 Apr;10(2):259-269. doi: 10.1007/s12551-017-0374-3. Epub 2017 Dec 14. Biophys Rev. 2018. PMID: 29243091 Free PMC article. Review.
-
Preferential assembly of heteromeric kainate and AMPA receptor amino terminal domains.Elife. 2017 Oct 23;6:e32056. doi: 10.7554/eLife.32056. Elife. 2017. PMID: 29058671 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

