Multiple G-quartet structures in pre-edited mRNAs suggest evolutionary driving force for RNA editing in trypanosomes
- PMID: 27436151
- PMCID: PMC4951716
- DOI: 10.1038/srep29810
Multiple G-quartet structures in pre-edited mRNAs suggest evolutionary driving force for RNA editing in trypanosomes
Abstract
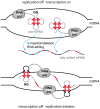
Mitochondrial transcript maturation in African trypanosomes requires a U-nucleotide specific RNA editing reaction. In its most extreme form hundreds of U's are inserted into and deleted from primary transcripts to generate functional mRNAs. Unfortunately, both origin and biological role of the process have remained enigmatic. Here we report a so far unrecognized structural feature of pre-edited mRNAs. We demonstrate that the cryptic pre-mRNAs contain numerous clustered G-nt, which fold into G-quadruplex (GQ) structures. We identified 27 GQ's in the different pre-mRNAs and demonstrate a positive correlation between the steady state abundance of guide (g)RNAs and the sequence position of GQ-elements. We postulate that the driving force for selecting G-rich sequences lies in the formation of DNA/RNA hybrid G-quadruplex (HQ) structures between the pre-edited transcripts and the non-template strands of mitochondrial DNA. HQ's are transcription termination/replication initiation sites and thus guarantee an unperturbed replication of the mt-genome. This is of special importance in the insect-stage of the parasite. In the transcription-on state, the identified GQ's require editing as a GQ-resolving activity indicating a link between replication, transcription and RNA editing. We propose that the different processes have coevolved and suggest the parasite life-cycle and the single mitochondrion as evolutionary driving forces.
Figures



References
-
- Kennedy P. G. E. Clinical features, diagnosis, and treatment of human African trypanosomiasis (sleeping sickness). Lancet Neurol. 12, 186–194 (2013). - PubMed
-
- Jensen R. E. & Englund P. T. Network news: the replication of kinetoplast DNA. Annu. Rev. Microbiol. 66, 473–491 (2012). - PubMed
-
- Benne R. et al. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46, 819–826 (1986). - PubMed
-
- Feagin J. E., Abraham J. M. & Stuart K. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell 53, 413–422 (1988). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

