Systems analysis of cis-regulatory motifs in C4 photosynthesis genes using maize and rice leaf transcriptomic data during a process of de-etiolation
- PMID: 27436282
- PMCID: PMC5014158
- DOI: 10.1093/jxb/erw275
Systems analysis of cis-regulatory motifs in C4 photosynthesis genes using maize and rice leaf transcriptomic data during a process of de-etiolation
Erratum in
-
Corrigendum.J Exp Bot. 2017 May 17;68(11):3035. doi: 10.1093/jxb/erw461. J Exp Bot. 2017. PMID: 28007949 Free PMC article. No abstract available.
Abstract
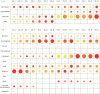
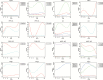
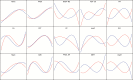
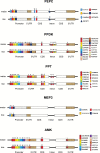
Identification of potential cis-regulatory motifs controlling the development of C4 photosynthesis is a major focus of current research. In this study, we used time-series RNA-seq data collected from etiolated maize and rice leaf tissues sampled during a de-etiolation process to systematically characterize the expression patterns of C4-related genes and to further identify potential cis elements in five different genomic regions (i.e. promoter, 5'UTR, 3'UTR, intron, and coding sequence) of C4 orthologous genes. The results demonstrate that although most of the C4 genes show similar expression patterns, a number of them, including chloroplast dicarboxylate transporter 1, aspartate aminotransferase, and triose phosphate transporter, show shifted expression patterns compared with their C3 counterparts. A number of conserved short DNA motifs between maize C4 genes and their rice orthologous genes were identified not only in the promoter, 5'UTR, 3'UTR, and coding sequences, but also in the introns of core C4 genes. We also identified cis-regulatory motifs that exist in maize C4 genes and also in genes showing similar expression patterns as maize C4 genes but that do not exist in rice C3 orthologs, suggesting a possible recruitment of pre-existing cis-elements from genes unrelated to C4 photosynthesis into C4 photosynthesis genes during C4 evolution.
Keywords: C4 photosynthesis; cell specificity; cis element; etiolation; evolution; systems biology..
© The Author 2016. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures

References
-
- Ali S, Taylor WC. 2001. Quantitative regulation of the Flaveria Me1 gene is controlled by the 3′-untranslated region and sequences near the amino terminus. Plant Molecular Biology 46, 251–261. - PubMed
-
- Aubry S, Brown NJ, Hibberd JM. 2011. The role of proteins in C3 plants prior to their recruitment into the C4 pathway. Journal of Experimental Botany 62, 3049–3059. - PubMed
-
- Aubry S, Smith-Unna RD, Boursnell CM, Kopriva S, Hibberd JM. 2014. Transcript residency on ribosomes reveals a key role for the Arabidopsis thaliana bundle sheath in sulfur and glucosinolate metabolism. Plant Journal 78, 659–673. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

