Length heterogeneity at conserved sequence block 2 in human mitochondrial DNA acts as a rheostat for RNA polymerase POLRMT activity
- PMID: 27436287
- PMCID: PMC5027508
- DOI: 10.1093/nar/gkw648
Length heterogeneity at conserved sequence block 2 in human mitochondrial DNA acts as a rheostat for RNA polymerase POLRMT activity
Abstract
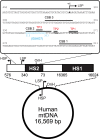
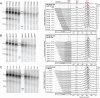
The guanine (G)-tract of conserved sequence block 2 (CSB 2) in human mitochondrial DNA can result in transcription termination due to formation of a hybrid G-quadruplex between the nascent RNA and the nontemplate DNA strand. This structure can then influence genome replication, stability and localization. Here we surveyed the frequency of variation in sequence identity and length at CSB 2 amongst human mitochondrial genomes and used in vitro transcription to assess the effects of this length heterogeneity on the activity of the mitochondrial RNA polymerase, POLRMT. In general, increased G-tract length correlated with increased termination levels. However, variation in the population favoured CSB 2 sequences which produced efficient termination while particularly weak or strong signals were avoided. For all variants examined, the 3' end of the transcripts mapped to the same downstream sequences and were prevented from terminating by addition of the transcription factor TEFM. We propose that CSB 2 length heterogeneity allows variation in the efficiency of transcription termination without affecting the position of the products or the capacity for regulation by TEFM.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
TEFM is a potent stimulator of mitochondrial transcription elongation in vitro.Nucleic Acids Res. 2015 Mar 11;43(5):2615-24. doi: 10.1093/nar/gkv105. Epub 2015 Feb 17. Nucleic Acids Res. 2015. PMID: 25690892 Free PMC article.
-
Mitochondrial biology. Replication-transcription switch in human mitochondria.Science. 2015 Jan 30;347(6221):548-51. doi: 10.1126/science.aaa0986. Science. 2015. PMID: 25635099 Free PMC article.
-
Mechanism of Transcription Anti-termination in Human Mitochondria.Cell. 2017 Nov 16;171(5):1082-1093.e13. doi: 10.1016/j.cell.2017.09.035. Epub 2017 Oct 12. Cell. 2017. PMID: 29033127 Free PMC article.
-
Human mitochondrial RNA polymerase: structure-function, mechanism and inhibition.Biochim Biophys Acta. 2012 Sep-Oct;1819(9-10):948-60. doi: 10.1016/j.bbagrm.2012.04.002. Epub 2012 Apr 19. Biochim Biophys Acta. 2012. PMID: 22551784 Review.
-
Nuclear gadgets in mitochondrial DNA replication and transcription.Trends Biochem Sci. 1991 Mar;16(3):107-11. doi: 10.1016/0968-0004(91)90043-u. Trends Biochem Sci. 1991. PMID: 2057998 Review.
Cited by
-
Involvement of the MGF 110-11L Gene in the African Swine Fever Replication and Virulence.Vaccines (Basel). 2023 Apr 14;11(4):846. doi: 10.3390/vaccines11040846. Vaccines (Basel). 2023. PMID: 37112759 Free PMC article.
-
Structural basis of mitochondrial transcription.Nat Struct Mol Biol. 2018 Sep;25(9):754-765. doi: 10.1038/s41594-018-0122-9. Epub 2018 Sep 6. Nat Struct Mol Biol. 2018. PMID: 30190598 Free PMC article. Review.
-
Mitochondrial DNA variation across 56,434 individuals in gnomAD.Genome Res. 2022 Mar;32(3):569-582. doi: 10.1101/gr.276013.121. Epub 2022 Jan 24. Genome Res. 2022. PMID: 35074858 Free PMC article.
-
Nuclear genetic control of mtDNA copy number and heteroplasmy in humans.Nature. 2023 Aug;620(7975):839-848. doi: 10.1038/s41586-023-06426-5. Epub 2023 Aug 16. Nature. 2023. PMID: 37587338 Free PMC article.
-
DNA specificities modulate the binding of human transcription factor A to mitochondrial DNA control region.Nucleic Acids Res. 2019 Jul 9;47(12):6519-6537. doi: 10.1093/nar/gkz406. Nucleic Acids Res. 2019. PMID: 31114891 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

