Toward high-resolution population genomics using archaeological samples
- PMID: 27436340
- PMCID: PMC4991838
- DOI: 10.1093/dnares/dsw029
Toward high-resolution population genomics using archaeological samples
Abstract
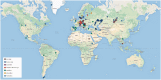
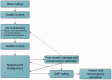
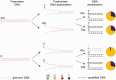
The term 'ancient DNA' (aDNA) is coming of age, with over 1,200 hits in the PubMed database, beginning in the early 1980s with the studies of 'molecular paleontology'. Rooted in cloning and limited sequencing of DNA from ancient remains during the pre-PCR era, the field has made incredible progress since the introduction of PCR and next-generation sequencing. Over the last decade, aDNA analysis ushered in a new era in genomics and became the method of choice for reconstructing the history of organisms, their biogeography, and migration routes, with applications in evolutionary biology, population genetics, archaeogenetics, paleo-epidemiology, and many other areas. This change was brought by development of new strategies for coping with the challenges in studying aDNA due to damage and fragmentation, scarce samples, significant historical gaps, and limited applicability of population genetics methods. In this review, we describe the state-of-the-art achievements in aDNA studies, with particular focus on human evolution and demographic history. We present the current experimental and theoretical procedures for handling and analysing highly degraded aDNA. We also review the challenges in the rapidly growing field of ancient epigenomics. Advancement of aDNA tools and methods signifies a new era in population genetics and evolutionary medicine research.
Keywords: ancient DNA; bioinformatics; epigenetics; next-generation sequencing; population genetics.
© The Author 2016. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Figures

Similar articles
-
Protocol for a comprehensive pipeline to study ancient human genomes.STAR Protoc. 2024 Jun 21;5(2):102985. doi: 10.1016/j.xpro.2024.102985. Epub 2024 Apr 30. STAR Protoc. 2024. PMID: 38691462 Free PMC article.
-
Human evolution: a tale from ancient genomes.Philos Trans R Soc Lond B Biol Sci. 2017 Feb 5;372(1713):20150484. doi: 10.1098/rstb.2015.0484. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 27994125 Free PMC article. Review.
-
Reconstructing DNA methylation maps of ancient populations.Nucleic Acids Res. 2024 Feb 28;52(4):1602-1612. doi: 10.1093/nar/gkad1232. Nucleic Acids Res. 2024. PMID: 38261973 Free PMC article.
-
Evolutionary anthropology and genes: investigating the genetics of human evolution from excavated skeletal remains.Gene. 2013 Oct 1;528(1):27-32. doi: 10.1016/j.gene.2013.06.011. Epub 2013 Jun 18. Gene. 2013. PMID: 23792014
-
Reinforcing plant evolutionary genomics using ancient DNA.Curr Opin Plant Biol. 2017 Apr;36:38-45. doi: 10.1016/j.pbi.2017.01.002. Epub 2017 Feb 1. Curr Opin Plant Biol. 2017. PMID: 28160617 Review.
Cited by
-
ARIADNA: machine learning method for ancient DNA variant discovery.DNA Res. 2018 Dec 1;25(6):619-627. doi: 10.1093/dnares/dsy029. DNA Res. 2018. PMID: 30215675 Free PMC article.
-
Advancements and Challenges in Ancient DNA Research: Bridging the Global North-South Divide.Genes (Basel). 2023 Feb 14;14(2):479. doi: 10.3390/genes14020479. Genes (Basel). 2023. PMID: 36833406 Free PMC article. Review.
-
Ancient Ancestry Informative Markers for Identifying Fine-Scale Ancient Population Structure in Eurasians.Genes (Basel). 2018 Dec 12;9(12):625. doi: 10.3390/genes9120625. Genes (Basel). 2018. PMID: 30545160 Free PMC article.
-
Evolutionary adaptation revealed by comparative genome analysis of woolly mammoths and elephants.DNA Res. 2017 Aug 1;24(4):359-369. doi: 10.1093/dnares/dsx007. DNA Res. 2017. PMID: 28369217 Free PMC article.
-
aYChr-DB: a database of ancient human Y haplogroups.NAR Genom Bioinform. 2020 Oct 9;2(4):lqaa081. doi: 10.1093/nargab/lqaa081. eCollection 2020 Dec. NAR Genom Bioinform. 2020. PMID: 33575627 Free PMC article.
References
-
- Orlando L., Ginolhac A., Zhang G., et al. 2013, Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature, 499, 74–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

