Loss of mutL homolog-1 (MLH1) expression promotes acquisition of oncogenic and inhibitor-resistant point mutations in tyrosine kinases
- PMID: 27436342
- PMCID: PMC11108519
- DOI: 10.1007/s00018-016-2310-2
Loss of mutL homolog-1 (MLH1) expression promotes acquisition of oncogenic and inhibitor-resistant point mutations in tyrosine kinases
Abstract
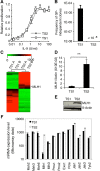
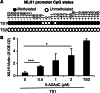
Genomic instability drives cancer progression by promoting genetic abnormalities that allow for the multi-step clonal selection of cells with growth advantages. We previously reported that the IL-9-dependent TS1 cell line sequentially acquired activating substitutions in JAK1 and JAK3 upon successive selections for growth factor independent and JAK inhibitor-resistant cells, suggestive of a defect in mutation avoidance mechanisms. In the first part of this paper, we discovered that the gene encoding mutL homolog-1 (MLH1), a key component of the DNA mismatch repair system, is silenced by promoter methylation in TS1 cells. By means of stable ectopic expression and RNA interference methods, we showed that the high frequencies of growth factor-independent and inhibitor-resistant cells with activating JAK mutations can be attributed to the absence of MLH1 expression. In the second part of this paper, we confirm the clinical relevance of our findings by showing that chronic myeloid leukemia relapses upon ABL-targeted therapy correlated with a lower expression of MLH1 messenger RNA. Interestingly, the mutational profile observed in our TS1 model, characterized by a strong predominance of T:A>C:G transitions, was identical to the one described in the literature for primitive cells derived from chronic myeloid leukemia patients. Taken together, our observations demonstrate for the first time a causal relationship between MLH1-deficiency and incidence of oncogenic point mutations in tyrosine kinases driving cell transformation and acquired resistance to kinase-targeted cancer therapies.
Keywords: DNA mismatch repair; Drug resistance; Janus kinase (JAK); MLH1; Oncogenic mutations; Tyrosine kinase inhibitor.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures





References
-
- Loeb LA. A mutator phenotype in cancer. Cancer Res. 2001;61(8):3230–3239. - PubMed
-
- Roche-Lestienne C, Soenen-Cornu V, Grardel-Duflos N, et al. Several types of mutations of the Abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood. 2002;100(3):1014–1018. doi: 10.1182/blood.V100.3.1014. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

