MEIOTIC F-BOX Is Essential for Male Meiotic DNA Double-Strand Break Repair in Rice
- PMID: 27436711
- PMCID: PMC5006700
- DOI: 10.1105/tpc.16.00108
MEIOTIC F-BOX Is Essential for Male Meiotic DNA Double-Strand Break Repair in Rice
Abstract
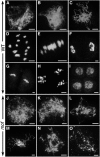
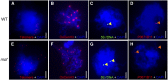
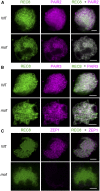
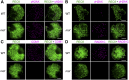
F-box proteins constitute a large superfamily in plants and play important roles in controlling many biological processes, but the roles of F-box proteins in male meiosis in plants remain unclear. Here, we identify the rice (Oryza sativa) F-box gene MEIOTIC F-BOX (MOF), which is essential for male meiotic progression. MOF belongs to the FBX subfamily and is predominantly active during leptotene to pachytene of prophase I. mof meiocytes display disrupted telomere bouquet formation, impaired pairing and synapsis of homologous chromosomes, and arrested meiocytes at late prophase I, followed by apoptosis. Although normal, programmed double-stranded DNA breaks (DSBs) form in mof mutants, foci of the phosphorylated histone variant γH2AX, a marker for DSBs, persist in the mutant, indicating that many of the DSBs remained unrepaired. The recruitment of Completion of meiosis I (COM1) and Radiation sensitive51C (RAD51C) to DSBs is severely compromised in mutant meiocytes, indicating that MOF is crucial for DSB end-processing and repair. Further analyses showed that MOF could physically interact with the rice SKP1-like Protein1 (OSK1), indicating that MOF functions as a component of the SCF E3 ligase to regulate meiotic progression in rice. Thus, this study reveals the essential role of an F-box protein in plant meiosis and provides helpful information for elucidating the roles of the ubiquitin proteasome system in plant meiotic progression.
© 2016 American Society of Plant Biologists. All rights reserved.
Figures


Similar articles
-
XRCC3 is essential for proper double-strand break repair and homologous recombination in rice meiosis.J Exp Bot. 2015 Sep;66(19):5713-25. doi: 10.1093/jxb/erv253. Epub 2015 Jun 1. J Exp Bot. 2015. PMID: 26034131
-
The F-Box Protein ZYGO1 Mediates Bouquet Formation to Promote Homologous Pairing, Synapsis, and Recombination in Rice Meiosis.Plant Cell. 2017 Oct;29(10):2597-2609. doi: 10.1105/tpc.17.00287. Epub 2017 Sep 22. Plant Cell. 2017. PMID: 28939596 Free PMC article.
-
The Exonuclease Homolog OsRAD1 Promotes Accurate Meiotic Double-Strand Break Repair by Suppressing Nonhomologous End Joining.Plant Physiol. 2016 Oct;172(2):1105-1116. doi: 10.1104/pp.16.00831. Epub 2016 Aug 10. Plant Physiol. 2016. PMID: 27512017 Free PMC article.
-
DNA double strand break repair, chromosome synapsis and transcriptional silencing in meiosis.Epigenetics. 2010 May 16;5(4):255-66. doi: 10.4161/epi.5.4.11518. Epub 2010 May 16. Epigenetics. 2010. PMID: 20364103 Review.
-
Double-stranded DNA breaks and gene functions in recombination and meiosis.Cell Res. 2006 May;16(5):402-12. doi: 10.1038/sj.cr.7310052. Cell Res. 2006. PMID: 16699536 Review.
Cited by
-
OsPRD2 is essential for double-strand break formation, but not spindle assembly during rice meiosis.Front Plant Sci. 2023 Jan 13;13:1122202. doi: 10.3389/fpls.2022.1122202. eCollection 2022. Front Plant Sci. 2023. PMID: 36714725 Free PMC article.
-
Cytological and proteomic analyses of floral buds reveal an altered atlas of meiosis in autopolyploid Brassica rapa.Cell Biosci. 2019 Jun 17;9:49. doi: 10.1186/s13578-019-0313-z. eCollection 2019. Cell Biosci. 2019. PMID: 31236208 Free PMC article.
-
Rice OsBRCA2 Is Required for DNA Double-Strand Break Repair in Meiotic Cells.Front Plant Sci. 2020 Nov 16;11:600820. doi: 10.3389/fpls.2020.600820. eCollection 2020. Front Plant Sci. 2020. PMID: 33304374 Free PMC article.
-
MORE FLORET1 Interacts with C-type Replication Protein A Complex and Regulates Male Meiosis in Rice.Rice (N Y). 2025 Apr 26;18(1):30. doi: 10.1186/s12284-025-00791-7. Rice (N Y). 2025. PMID: 40285806 Free PMC article.
-
Ubiquitination in Plant Meiosis: Recent Advances and High Throughput Methods.Front Plant Sci. 2021 Apr 7;12:667314. doi: 10.3389/fpls.2021.667314. eCollection 2021. Front Plant Sci. 2021. PMID: 33897750 Free PMC article. Review.
References
-
- Abdu U., Brodsky M., Schüpbach T. (2002). Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of Gurken through Chk2/Mnk. Curr. Biol. 12: 1645–1651. - PubMed
-
- Abe A., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30: 174–178. - PubMed
-
- Bergerat A., de Massy B., Gadelle D., Varoutas P.C., Nicolas A., Forterre P. (1997). An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386: 414–417. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

