Indel Group in Genomes (IGG) Molecular Genetic Markers
- PMID: 27436831
- PMCID: PMC5074621
- DOI: 10.1104/pp.16.00354
Indel Group in Genomes (IGG) Molecular Genetic Markers
Abstract
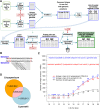
Genetic markers are essential when developing or working with genetically variable populations. Indel Group in Genomes (IGG) markers are primer pairs that amplify single-locus sequences that differ in size for two or more alleles. They are attractive for their ease of use for rapid genotyping and their codominant nature. Here, we describe a heuristic algorithm that uses a k-mer-based approach to search two or more genome sequences to locate polymorphic regions suitable for designing candidate IGG marker primers. As input to the IGG pipeline software, the user provides genome sequences and the desired amplicon sizes and size differences. Primer sequences flanking polymorphic insertions/deletions are produced as output. IGG marker files for three sets of genomes, Solanum lycopersicum/Solanum pennellii, Arabidopsis (Arabidopsis thaliana) Columbia-0/Landsberg erecta-0 accessions, and S. lycopersicum/S. pennellii/Solanum tuberosum (three-way polymorphic) are included.
© 2016 American Society of Plant Biologists. All rights reserved.
Figures



References
-
- Ahmed A, Ferreira AS, Hartskeerl RA (2015) Multilocus sequence typing (MLST): markers for the traceability of pathogenic Leptospira strains. Methods Mol Biol 1247: 349–359 - PubMed
-
- Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA (2011) The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res 39: D1149–D1155 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

