Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers
- PMID: 27436900
- PMCID: PMC4995967
- DOI: 10.1073/pnas.1609060113
Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers
Abstract
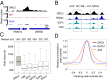
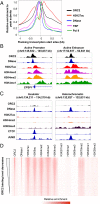
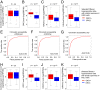
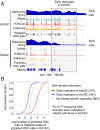
The origin recognition complex (ORC) binds sites from which DNA replication is initiated. We address ORC binding selectivity in vivo by mapping ∼52,000 ORC2 binding sites throughout the human genome. The ORC binding profile is broader than those of sequence-specific transcription factors, suggesting that ORC is not bound or recruited to specific DNA sequences. Instead, ORC binds nonspecifically to open (DNase I-hypersensitive) regions containing active chromatin marks such as H3 acetylation and H3K4 methylation. ORC sites in early and late replicating regions have similar properties, but there are far more ORC sites in early replicating regions. This suggests that replication timing is due primarily to ORC density and stochastic firing of origins. Computational simulation of stochastic firing from identified ORC sites is in accord with replication timing data. Large genomic regions with a paucity of ORC sites are strongly associated with common fragile sites and recurrent deletions in cancers. We suggest that replication origins, replication timing, and replication-dependent chromosome breaks are determined primarily by the genomic distribution of activator proteins at enhancers and promoters. These activators recruit nucleosome-modifying complexes to create the appropriate chromatin structure that allows ORC binding and subsequent origin firing.
Keywords: DNA replication; ORC; chromatin; replication origins; replication timing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
- 9136 doi: 10.1073/pnas.1610336113
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

