A representation of a compressed de Bruijn graph for pan-genome analysis that enables search
- PMID: 27437028
- PMCID: PMC4950428
- DOI: 10.1186/s13015-016-0083-7
A representation of a compressed de Bruijn graph for pan-genome analysis that enables search
Erratum in
-
Erratum to: A representation of a compressed de Bruijn graph for pan-genome analysis that enables search.Algorithms Mol Biol. 2016 Nov 28;11:28. doi: 10.1186/s13015-016-0090-8. eCollection 2016. Algorithms Mol Biol. 2016. PMID: 27933096 Free PMC article.
Abstract
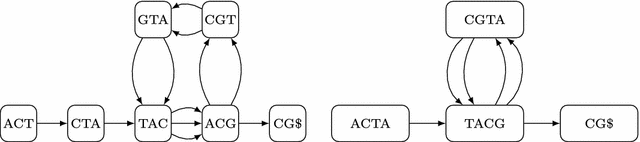
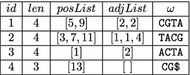
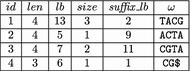
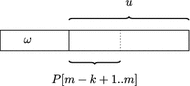
Background: Recently, Marcus et al. (Bioinformatics 30:3476-83, 2014) proposed to use a compressed de Bruijn graph to describe the relationship between the genomes of many individuals/strains of the same or closely related species. They devised an [Formula: see text] time algorithm called splitMEM that constructs this graph directly (i.e., without using the uncompressed de Bruijn graph) based on a suffix tree, where n is the total length of the genomes and g is the length of the longest genome. Baier et al. (Bioinformatics 32:497-504, 2016) improved their result.
Results: In this paper, we propose a new space-efficient representation of the compressed de Bruijn graph that adds the possibility to search for a pattern (e.g. an allele-a variant form of a gene) within the pan-genome. The ability to search within the pan-genome graph is of utmost importance and is a design goal of pan-genome data structures.
Keywords: Backward search; Burrows–Wheeler transform; Compressed de Bruijn graph; Pan-genome analysis.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

