Relationship between Metabolic Fluxes and Sequence-Derived Properties of Enzymes
- PMID: 27437461
- PMCID: PMC4897147
- DOI: 10.1155/2014/817102
Relationship between Metabolic Fluxes and Sequence-Derived Properties of Enzymes
Abstract
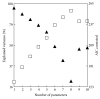
Metabolic fluxes are key parameters of metabolic pathways being closely related to the kinetic properties of enzymes, thereby could be dependent on. This study examines possible relationships between the metabolic fluxes and the physical-chemical/structural features of enzymes from the yeast Saccharomyces cerevisiae glycolysis pathway. Metabolic fluxes were quantified by the COPASI tool using the kinetic models of Hynne and Teusink at varied concentrations of external glucose. The enzyme sequences were taken from the UniProtKB and the average amino acid (AA) properties were computed using the set of Georgiev's uncorrelated scales that satisfy the VARIMAX criterion and specific AA indices that show the highest correlations with those. Multiple linear regressions (88.41% <R adjusted (2) < 93.32%; P < 0.00001) were found between the values of metabolic fluxes and the selected sets of the average AA properties. The hydrophobicity, α-helicity, and net charge were pointed out as the most influential characteristics of the sequences. The results provide an evidence that metabolic fluxes of the yeast glycolysis pathway are closely related to certain physical-chemical properties of relevant enzymes and support the view on the interdependence of catalytic, binding, and structural AA residues to ensure the efficiency of biocatalysts and, hence, physiologically adequate metabolic processes.
Figures


Similar articles
-
Relationships between kinetic constants and the amino acid composition of enzymes from the yeast Saccharomyces cerevisiae glycolysis pathway.EURASIP J Bioinform Syst Biol. 2012 Aug 6;2012(1):11. doi: 10.1186/1687-4153-2012-11. EURASIP J Bioinform Syst Biol. 2012. PMID: 22867018 Free PMC article.
-
Continuous modeling of metabolic networks with gene regulation in yeast and in vivo determination of rate parameters.Biotechnol Bioeng. 2012 Sep;109(9):2325-39. doi: 10.1002/bit.24503. Epub 2012 Apr 24. Biotechnol Bioeng. 2012. PMID: 22447363
-
Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry.Eur J Biochem. 2000 Sep;267(17):5313-29. doi: 10.1046/j.1432-1327.2000.01527.x. Eur J Biochem. 2000. PMID: 10951190
-
'Slave' metabolites and enzymes. A rapid way of delineating metabolic control.Eur J Biochem. 2000 Apr;267(7):1889-93. doi: 10.1046/j.1432-1327.2000.01220.x. Eur J Biochem. 2000. PMID: 10727927 Review.
-
The importance of post-translational modifications in regulating Saccharomyces cerevisiae metabolism.FEMS Yeast Res. 2012 Mar;12(2):104-17. doi: 10.1111/j.1567-1364.2011.00765.x. Epub 2011 Dec 22. FEMS Yeast Res. 2012. PMID: 22128902 Review.
References
-
- Plaxton W. Functional Metabolism: Regulation and Adaptation. New York, NY, USA: Wiley- Liss; 2004. Principles of metabolic control; pp. 1–24.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

