Whole Genome Sequencing Identifies a 78 kb Insertion from Chromosome 8 as the Cause of Charcot-Marie-Tooth Neuropathy CMTX3
- PMID: 27438001
- PMCID: PMC4954712
- DOI: 10.1371/journal.pgen.1006177
Whole Genome Sequencing Identifies a 78 kb Insertion from Chromosome 8 as the Cause of Charcot-Marie-Tooth Neuropathy CMTX3
Abstract
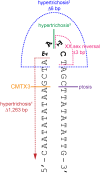
With the advent of whole exome sequencing, cases where no pathogenic coding mutations can be found are increasingly being observed in many diseases. In two large, distantly-related families that mapped to the Charcot-Marie-Tooth neuropathy CMTX3 locus at chromosome Xq26.3-q27.3, all coding mutations were excluded. Using whole genome sequencing we found a large DNA interchromosomal insertion within the CMTX3 locus. The 78 kb insertion originates from chromosome 8q24.3, segregates fully with the disease in the two families, and is absent from the general population as well as 627 neurologically normal chromosomes from in-house controls. Large insertions into chromosome Xq27.1 are known to cause a range of diseases and this is the first neuropathy phenotype caused by an interchromosomal insertion at this locus. The CMTX3 insertion represents an understudied pathogenic structural variation mechanism for inherited peripheral neuropathies. Our finding highlights the importance of considering all structural variation types when studying unsolved inherited peripheral neuropathy cases with no pathogenic coding mutations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

