Applications of (19)F-NMR in Fragment-Based Drug Discovery
- PMID: 27438818
- PMCID: PMC6273323
- DOI: 10.3390/molecules21070860
Applications of (19)F-NMR in Fragment-Based Drug Discovery
Abstract
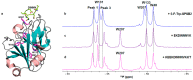
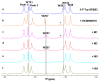
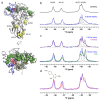
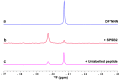
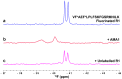
(19)F-NMR has proved to be a valuable tool in fragment-based drug discovery. Its applications include screening libraries of fluorinated fragments, assessing competition among elaborated fragments and identifying the binding poses of promising hits. By observing fluorine in both the ligand and the target protein, useful information can be obtained on not only the binding pose but also the dynamics of ligand-protein interactions. These applications of (19)F-NMR will be illustrated in this review with studies from our fragment-based drug discovery campaigns against protein targets in parasitic and infectious diseases.
Keywords: 19F-NMR; chemical shift; fragment-based drug design; labelling; ligand; linewidth; peptide; protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

