TALE-directed local modulation of H3K9 methylation shapes exon recognition
- PMID: 27439481
- PMCID: PMC4954949
- DOI: 10.1038/srep29961
TALE-directed local modulation of H3K9 methylation shapes exon recognition
Abstract
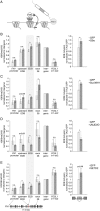
In search for the function of local chromatin environment on pre-mRNA processing we established a new tool, which allows for the modification of chromatin using a targeted approach. Using Transcription Activator-Like Effector domains fused to histone modifying enzymes (TALE-HME), we show locally restricted alteration of histone methylation modulates the splicing of target exons. We provide evidence that a local increase in H3K9 di- and trimethylation promotes inclusion of the target alternative exon, while demethylation by JMJD2D leads to exon skipping. We further demonstrate that H3K9me3 is localized on internal exons genome-wide suggesting a general role in splicing. Consistently, targeting of the H3K9 demethylase to a weak constitutive exon reduced co-transcriptional splicing. Together our data show H3K9 methylation within the gene body is a factor influencing recognition of both constitutive and alternative exons.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

