Concurrence of Iridovirus, Polyomavirus, and a Unique Member of a New Group of Fish Papillomaviruses in Lymphocystis Disease-Affected Gilthead Sea Bream
- PMID: 27440877
- PMCID: PMC5021401
- DOI: 10.1128/JVI.01369-16
Concurrence of Iridovirus, Polyomavirus, and a Unique Member of a New Group of Fish Papillomaviruses in Lymphocystis Disease-Affected Gilthead Sea Bream
Abstract
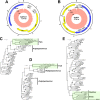
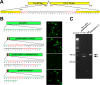
Lymphocystis disease is a geographically widespread disease affecting more than 150 different species of marine and freshwater fish. The disease, provoked by the iridovirus lymphocystis disease virus (LCDV), is characterized by the appearance of papillomalike lesions on the skin of affected animals that usually self-resolve over time. Development of the disease is usually associated with several environmental factors and, more frequently, with stress conditions provoked by the intensive culture conditions present in fish farms. In gilthead sea bream (Sparus aurata), an economically important cultured fish species in the Mediterranean area, a distinct LCDV has been identified but not yet completely characterized. We have used direct sequencing of the virome of lymphocystis lesions from affected S. aurata fish to obtain the complete genome of a new LCDV-Sa species that is the largest vertebrate iridovirus sequenced to date. Importantly, this approach allowed us to assemble the full-length circular genome sequence of two previously unknown viruses belonging to the papillomaviruses and polyomaviruses, termed Sparus aurata papillomavirus 1 (SaPV1) and Sparus aurata polyomavirus 1 (SaPyV1), respectively. Epidemiological surveys showed that lymphocystis disease was frequently associated with the concurrent appearance of one or both of the new viruses. SaPV1 has unique characteristics, such as an intron within the L1 gene, and as the first member of the Papillomaviridae family described in fish, provides evidence for a more ancient origin of this family than previously thought.
Importance: Lymphocystis disease affects marine and freshwater fish species worldwide. It is characterized by the appearance of papillomalike lesions on the skin that contain heavily enlarged cells (lymphocysts). The causative agent is the lymphocystis disease virus (LCDV), a large icosahedral virus of the family Iridoviridae In the Mediterranean area, the gilthead sea bream (Sparus aurata), an important farmed fish, is frequently affected. Using next-generation sequencing, we have identified within S. aurata lymphocystis lesions the concurrent presence of an additional LCDV species (LCDV-Sa) as well as two novel viruses. These are members of polyomavirus and papillomavirus families, and here we report them to be frequently associated with the presence of lymphocysts in affected fish. Because papillomaviruses have not been described in fish before, these findings support a more ancient origin of this virus family than previously thought and evolutionary implications are discussed.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Smail DA, Munro ES. 2012. The virology of teleosts, p 186–291. In Roberts RJ. (ed), Fish pathology, 4th ed, Wiley-Blackwell, Oxford, United Kingdom.
-
- Weissenberg R. 1949. Studies on lymphocystis tumor cells of fish; the osmiophilic granules of the cytoplasmic inclusions and their interpretation as elementary bodies of the lymphocystis virus. Cancer Res 9:537–542. - PubMed
-
- Weissenberg R. 1914. Über infectiöse Zellhypertrophie bei Fischen (Lymphocystiserkrankung). Sitzungsber Kgl Preuss Akad Wiss Sitz Physik Mathem 16:792.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

