Viral Macro Domains Reverse Protein ADP-Ribosylation
- PMID: 27440879
- PMCID: PMC5021415
- DOI: 10.1128/JVI.00705-16
Viral Macro Domains Reverse Protein ADP-Ribosylation
Abstract
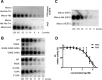
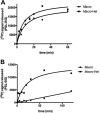
ADP-ribosylation is a posttranslational protein modification in which ADP-ribose is transferred from NAD(+) to specific acceptors to regulate a wide variety of cellular processes. The macro domain is an ancient and highly evolutionarily conserved protein domain widely distributed throughout all kingdoms of life, including viruses. The human TARG1/C6orf130, MacroD1, and MacroD2 proteins can reverse ADP-ribosylation by acting on ADP-ribosylated substrates through the hydrolytic activity of their macro domains. Here, we report that the macro domain from hepatitis E virus (HEV) serves as an ADP-ribose-protein hydrolase for mono-ADP-ribose (MAR) and poly(ADP-ribose) (PAR) chain removal (de-MARylation and de-PARylation, respectively) from mono- and poly(ADP)-ribosylated proteins, respectively. The presence of the HEV helicase in cis dramatically increases the binding of the macro domain to poly(ADP-ribose) and stimulates the de-PARylation activity. Abrogation of the latter dramatically decreases replication of an HEV subgenomic replicon. The de-MARylation activity is present in all three pathogenic positive-sense, single-stranded RNA [(+)ssRNA] virus families which carry a macro domain: Coronaviridae (severe acute respiratory syndrome coronavirus and human coronavirus 229E), Togaviridae (Venezuelan equine encephalitis virus), and Hepeviridae (HEV), indicating that it might be a significant tropism and/or pathogenic determinant.
Importance: Protein ADP-ribosylation is a covalent posttranslational modification regulating cellular protein activities in a dynamic fashion to modulate and coordinate a variety of cellular processes. Three viral families, Coronaviridae, Togaviridae, and Hepeviridae, possess macro domains embedded in their polyproteins. Here, we show that viral macro domains reverse cellular ADP-ribosylation, potentially cutting the signal of a viral infection in the cell. Various poly(ADP-ribose) polymerases which are notorious guardians of cellular integrity are demodified by macro domains from members of these virus families. In the case of hepatitis E virus, the adjacent viral helicase domain dramatically increases the binding of the macro domain to PAR and simulates the demodification activity.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures



References
-
- Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, Simpson MA, Matic I, Ozkan E, Golia B, Schellenberg MJ, Weston R, Williams JG, Rossi MN, Galehdari H, Krahn J, Wan A, Trembath RC, Crosby AH, Ahel D, Hay R, Ladurner AG, Timinszky G, Williams RS, Ahel I. 2013. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J 32:1225–1237. doi:10.1038/emboj.2013.51. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

