Hepatitis B Virus Polymerase Localizes to the Mitochondria, and Its Terminal Protein Domain Contains the Mitochondrial Targeting Signal
- PMID: 27440888
- PMCID: PMC5021430
- DOI: 10.1128/JVI.01229-16
Hepatitis B Virus Polymerase Localizes to the Mitochondria, and Its Terminal Protein Domain Contains the Mitochondrial Targeting Signal
Abstract
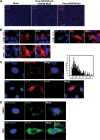
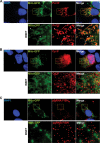
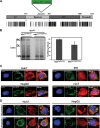
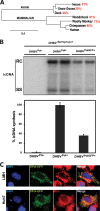
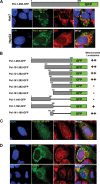
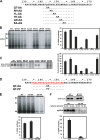
To understand subcellular sites of hepatitis B virus (HBV) replication, we visualized core (Cp), polymerase (Pol), and pregenomic RNA (pgRNA) in infected cells. Interestingly, we found that the majority of Pol localized to the mitochondria in cells undergoing viral replication. The mitochondrial localization of Pol was independent of both the cell type and other viral components, indicating that Pol contains an intrinsic mitochondrial targeting signal (MTS). Neither Cp nor pgRNA localized to the mitochondria during active replication, suggesting a role other than DNA synthesis for Pol at the mitochondria. The Pol of duck hepatitis B virus (DHBV) also localized to the mitochondria. This result indicates that localization of Pol to mitochondria is likely a feature of all hepadnaviruses. To map the MTS within HBV Pol, we generated a series of Pol-green fluorescent protein (Pol-GFP) fusions and found that a stretch spanning amino acids (aa) 141 to 160 of Pol was sufficient to target GFP to the mitochondria. Surprisingly, deleting aa 141 to 160 in full-length Pol did not fully ablate Pol's mitochondrial localization, suggesting that additional sequences are involved in mitochondrial targeting. Only by deleting the N-terminal 160 amino acids in full-length Pol was mitochondrial localization ablated. Crucial residues for pgRNA packaging are contained within aa 141 to 160, indicating a multifunctional role of this region of Pol in the viral life cycle. Our studies show an unexpected Pol trafficking behavior that is uncoupled from its role in viral DNA synthesis.
Importance: Chronic infection by HBV is a serious health concern. Existing therapies for chronically infected individuals are not curative, underscoring the need for a better understanding of the viral life cycle to develop better antiviral therapies. To date, the most thoroughly studied function of Pol is to package the pgRNA and reverse transcribe it to double-stranded DNA within capsids. This study provides evidence for mitochondrial localization of Pol and defines the MTS. Recent findings have implicated a non-reverse transcription role for Pol in evading host innate immune responses. Mitochondria play an important role in controlling cellular metabolism, apoptosis, and innate immunity. Pol may alter one or more of these host mitochondrial functions to gain a replicative advantage and persist in chronically infected individuals.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
References
-
- Beasley RP. 1988. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer 61:1942–1956. - PubMed
-
- GBD 2013 Mortality and Causes of Death Collaborators. 2015. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 385:117–171. doi:10.1016/S0140-6736(14)61682-2. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

