In Silico Design and Experimental Validation of siRNAs Targeting Conserved Regions of Multiple Hepatitis C Virus Genotypes
- PMID: 27441640
- PMCID: PMC4956106
- DOI: 10.1371/journal.pone.0159211
In Silico Design and Experimental Validation of siRNAs Targeting Conserved Regions of Multiple Hepatitis C Virus Genotypes
Abstract
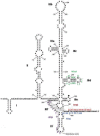
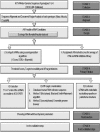
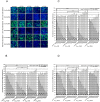
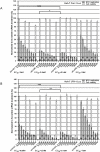
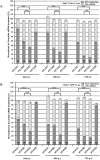
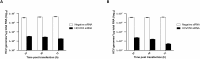
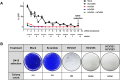
RNA interference (RNAi) is a post-transcriptional gene silencing mechanism that mediates the sequence-specific degradation of targeted RNA and thus provides a tremendous opportunity for development of oligonucleotide-based drugs. Here, we report on the design and validation of small interfering RNAs (siRNAs) targeting highly conserved regions of the hepatitis C virus (HCV) genome. To aim for therapeutic applications by optimizing the RNAi efficacy and reducing potential side effects, we considered different factors such as target RNA variations, thermodynamics and accessibility of the siRNA and target RNA, and off-target effects. This aim was achieved using an in silico design and selection protocol complemented by an automated MysiRNA-Designer pipeline. The protocol included the design and filtration of siRNAs targeting highly conserved and accessible regions within the HCV internal ribosome entry site, and adjacent core sequences of the viral genome with high-ranking efficacy scores. Off-target analysis excluded siRNAs with potential binding to human mRNAs. Under this strict selection process, two siRNAs (HCV353 and HCV258) were selected based on their predicted high specificity and potency. These siRNAs were tested for antiviral efficacy in HCV genotype 1 and 2 replicon cell lines. Both in silico-designed siRNAs efficiently inhibited HCV RNA replication, even at low concentrations and for short exposure times (24h); they also exceeded the antiviral potencies of reference siRNAs targeting HCV. Furthermore, HCV353 and HCV258 siRNAs also inhibited replication of patient-derived HCV genotype 4 isolates in infected Huh-7 cells. Prolonged treatment of HCV replicon cells with HCV353 did not result in the appearance of escape mutant viruses. Taken together, these results reveal the accuracy and strength of our integrated siRNA design and selection protocols. These protocols could be used to design highly potent and specific RNAi-based therapeutic oligonucleotide interventions.
Conflict of interest statement
Figures
Similar articles
-
Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs.J Virol. 2004 Apr;78(7):3436-46. doi: 10.1128/jvi.78.7.3436-3446.2004. J Virol. 2004. PMID: 15016866 Free PMC article.
-
Down-regulation of IRES containing 5'UTR of HCV genotype 3a using siRNAs.Virol J. 2011 May 13;8:221. doi: 10.1186/1743-422X-8-221. Virol J. 2011. PMID: 21569449 Free PMC article.
-
Inhibition of Hepatitis C Virus in Mice by a Small Interfering RNA Targeting a Highly Conserved Sequence in Viral IRES Pseudoknot.PLoS One. 2016 Jan 11;11(1):e0146710. doi: 10.1371/journal.pone.0146710. eCollection 2016. PLoS One. 2016. PMID: 26751678 Free PMC article.
-
Interfering with hepatitis C virus RNA replication.Virus Res. 2004 Jun 1;102(1):19-25. doi: 10.1016/j.virusres.2004.01.011. Virus Res. 2004. PMID: 15068876 Review.
-
[Replication of hepatitis C virus genome].Uirusu. 2008 Dec;58(2):191-8. Uirusu. 2008. PMID: 19374197 Review. Japanese.
Cited by
-
Bioinformatics analysis reveals four major hexon variants of human adenovirus type-3 (HAdV-3) as the potential strains for development of vaccine and siRNA-based therapeutics against HAdV-3 respiratory infections.Infect Genet Evol. 2020 Nov;85:104439. doi: 10.1016/j.meegid.2020.104439. Epub 2020 Jun 23. Infect Genet Evol. 2020. PMID: 32585339 Free PMC article.
-
Computational study and design of effective siRNAs to silence structural proteins associated genes of Indian SARS-CoV-2 strains.Comput Biol Chem. 2022 Jun;98:107687. doi: 10.1016/j.compbiolchem.2022.107687. Epub 2022 Apr 29. Comput Biol Chem. 2022. PMID: 35537364 Free PMC article.
-
In silico prediction and experimental validation of siRNAs targeting ORF1ab of MERS-CoV in Vero cell line.Saudi J Biol Sci. 2021 Feb;28(2):1348-1355. doi: 10.1016/j.sjbs.2020.11.066. Epub 2020 Nov 27. Saudi J Biol Sci. 2021. PMID: 33519276 Free PMC article.
-
Delivery of siRNAs against MERS-CoV in Vero and HEK-293 cells: A comparative evaluation of transfection reagents.J King Saud Univ Sci. 2023 Apr;35(3):102540. doi: 10.1016/j.jksus.2023.102540. Epub 2023 Jan 5. J King Saud Univ Sci. 2023. PMID: 36624781 Free PMC article.
-
Development, Design, and Application of Efficient siRNAs Against Cotton Leaf Curl Virus-Betasatellite Complex to Mediate Resistance Against Cotton Leaf Curl Disease.Indian J Microbiol. 2024 Jun;64(2):558-571. doi: 10.1007/s12088-024-01191-z. Epub 2024 Feb 3. Indian J Microbiol. 2024. PMID: 39011016 Free PMC article.
References
-
- Lindenbach BD, Rice CM (2005) Unravelling hepatitis C virus replication from genome to function. Nature 436: 933–938. - PubMed
-
- Van Regenmortel MH (2007) Virus species and virus identification: past and current controversies. Infect Genet Evol 7: 133–144. - PubMed
-
- Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, Feinstone S, et al. (2005) Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42: 962–973. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

