Regulation of Aerobic Energy Metabolism in Podospora anserina by Two Paralogous Genes Encoding Structurally Different c-Subunits of ATP Synthase
- PMID: 27442014
- PMCID: PMC4956034
- DOI: 10.1371/journal.pgen.1006161
Regulation of Aerobic Energy Metabolism in Podospora anserina by Two Paralogous Genes Encoding Structurally Different c-Subunits of ATP Synthase
Abstract
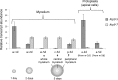
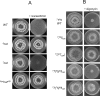
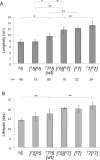
Most of the ATP in living cells is produced by an F-type ATP synthase. This enzyme uses the energy of a transmembrane electrochemical proton gradient to synthesize ATP from ADP and inorganic phosphate. Proton movements across the membrane domain (FO) of the ATP synthase drive the rotation of a ring of 8-15 c-subunits, which induces conformational changes in the catalytic part (F1) of the enzyme that ultimately promote ATP synthesis. Two paralogous nuclear genes, called Atp9-5 and Atp9-7, encode structurally different c-subunits in the filamentous fungus Podospora anserina. We have in this study identified differences in the expression pattern for the two genes that correlate with the mitotic activity of cells in vegetative mycelia: Atp9-7 is transcriptionally active in non-proliferating (stationary) cells while Atp9-5 is expressed in the cells at the extremity (apex) of filaments that divide and are responsible for mycelium growth. When active, the Atp9-5 gene sustains a much higher rate of c-subunit synthesis than Atp9-7. We further show that the ATP9-7 and ATP9-5 proteins have antagonist effects on the longevity of P. anserina. Finally, we provide evidence that the ATP9-5 protein sustains a higher rate of mitochondrial ATP synthesis and yield in ATP molecules per electron transferred to oxygen than the c-subunit encoded by Atp9-7. These findings reveal that the c-subunit genes play a key role in the modulation of ATP synthase production and activity along the life cycle of P. anserina. Such a degree of sophistication for regulating aerobic energy metabolism has not been described before.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Two nuclear life cycle-regulated genes encode interchangeable subunits c of mitochondrial ATP synthase in Podospora anserina.Mol Biol Evol. 2011 Jul;28(7):2063-75. doi: 10.1093/molbev/msr025. Epub 2011 Jan 27. Mol Biol Evol. 2011. PMID: 21273631
-
Experimental relocation of the mitochondrial ATP9 gene to the nucleus reveals forces underlying mitochondrial genome evolution.PLoS Genet. 2012;8(8):e1002876. doi: 10.1371/journal.pgen.1002876. Epub 2012 Aug 16. PLoS Genet. 2012. PMID: 22916027 Free PMC article.
-
Thiol oxidation of mitochondrial F0-c subunits: a way to switch off antimicrobial drug targets of the mitochondrial ATP synthase.Med Hypotheses. 2014 Aug;83(2):160-5. doi: 10.1016/j.mehy.2014.05.004. Epub 2014 May 17. Med Hypotheses. 2014. PMID: 24932580
-
Linking structural features from mitochondrial and bacterial F-type ATP synthases to their distinct mechanisms of ATPase inhibition.Prog Biophys Mol Biol. 2015 Oct;119(1):94-102. doi: 10.1016/j.pbiomolbio.2015.06.005. Epub 2015 Jul 2. Prog Biophys Mol Biol. 2015. PMID: 26140992 Review.
-
The c-Ring of the F1FO-ATP Synthase: Facts and Perspectives.J Membr Biol. 2016 Apr;249(1-2):11-21. doi: 10.1007/s00232-015-9860-3. Epub 2015 Nov 30. J Membr Biol. 2016. PMID: 26621635 Review.
Cited by
-
The Mitogenomes of Ophiostoma minus and Ophiostoma piliferum and Comparisons With Other Members of the Ophiostomatales.Front Microbiol. 2021 Feb 10;12:618649. doi: 10.3389/fmicb.2021.618649. eCollection 2021. Front Microbiol. 2021. PMID: 33643245 Free PMC article.
-
Characterization of the mitochondrial genomes for Ophiostomaips and related taxa from various geographic origins and related species: large intron-rich genomes and complex intron arrangements.IMA Fungus. 2025 Jul 22;16:e159349. doi: 10.3897/imafungus.16.159349. eCollection 2025. IMA Fungus. 2025. PMID: 40741210 Free PMC article.
-
The mitogenomes of Leptographium aureum, Leptographium sp., and Grosmannia fruticeta: expansion by introns.Front Microbiol. 2023 Aug 10;14:1240407. doi: 10.3389/fmicb.2023.1240407. eCollection 2023. Front Microbiol. 2023. PMID: 37637121 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

