Impact of structural polymorphism for the Helicobacter pylori CagA oncoprotein on binding to polarity-regulating kinase PAR1b
- PMID: 27445265
- PMCID: PMC4957108
- DOI: 10.1038/srep30031
Impact of structural polymorphism for the Helicobacter pylori CagA oncoprotein on binding to polarity-regulating kinase PAR1b
Abstract
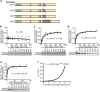
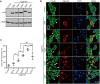
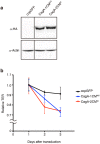
Chronic infection with cagA-positive Helicobacter pylori is the strongest risk factor for atrophic gastritis, peptic ulcers, and gastric cancer. CagA, the product of the cagA gene, is a bacterial oncoprotein, which, upon delivery into gastric epithelial cells, binds to and inhibits the polarity-regulating kinase, partitioning-defective 1b (PAR1b) [also known as microtubule affinity-regulating kinase 2 (MARK2)], via its CagA multimerization (CM) motif. The inhibition of PAR1b elicits junctional and polarity defects, rendering cells susceptible to oncogenesis. Notably, the polymorphism in the CM motif has been identified among geographic variants of CagA, differing in either the copy number or the sequence composition. In this study, through quantitative analysis of the complex formation between CagA and PAR1b, we found that several CagA species have acquired elevated PAR1b-binding activity via duplication of the CM motifs, while others have lost their PAR1b-binding activity. We also found that strength of CagA-PAR1b interaction was proportional to the degrees of stress fiber formation and tight junctional disruption by CagA in gastric epithelial cells. These results indicate that the CM polymorphism is a determinant for the magnitude of CagA-mediated deregulation of the cytoskeletal system and thereby possibly affects disease outcome of cagA-positive H. pylori infection, including gastric cancer.
Figures



References
-
- Peek R. M. & Blaser M. J. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2, 28–37 (2002). - PubMed
-
- Hatakeyama M. Helicobacter pylori CagA and gastric cancer: A paradigm for hit-and-run carcinogenesis. Cell Host Microbe 15, 306–316 (2014). - PubMed
-
- Nomura A. et al. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N. Engl. J. Med. 325, 1132–1136 (1991). - PubMed
-
- Parsonnet J. et al. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325, 1127–1131 (1991). - PubMed
-
- Ferlay J. et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386 (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

