Functional Diversity of Cytotoxic tRNase/Immunity Protein Complexes from Burkholderia pseudomallei
- PMID: 27445337
- PMCID: PMC5016678
- DOI: 10.1074/jbc.M116.736074
Functional Diversity of Cytotoxic tRNase/Immunity Protein Complexes from Burkholderia pseudomallei
Abstract
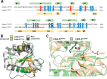
Contact-dependent growth inhibition (CDI) is a widespread mechanism of inter-bacterial competition. CDI(+) bacteria deploy large CdiA effector proteins, which carry variable C-terminal toxin domains (CdiA-CT). CDI(+) cells also produce CdiI immunity proteins that specifically neutralize cognate CdiA-CT toxins to prevent auto-inhibition. Here, we present the crystal structure of the CdiA-CT/CdiI(E479) toxin/immunity protein complex from Burkholderia pseudomallei isolate E479. The CdiA-CT(E479) tRNase domain contains a core α/β-fold that is characteristic of PD(D/E)XK superfamily nucleases. Unexpectedly, the closest structural homolog of CdiA-CT(E479) is another CDI toxin domain from B. pseudomallei 1026b. Although unrelated in sequence, the two B. pseudomallei nuclease domains share similar folds and active-site architectures. By contrast, the CdiI(E479) and CdiI(1026b) immunity proteins share no significant sequence or structural homology. CdiA-CT(E479) and CdiA-CT(1026b) are both tRNases; however, each nuclease cleaves tRNA at a distinct position. We used a molecular docking approach to model each toxin bound to tRNA substrate. The resulting models fit into electron density envelopes generated by small-angle x-ray scattering analysis of catalytically inactive toxin domains bound stably to tRNA. CdiA-CT(E479) is the third CDI toxin found to have structural homology to the PD(D/E)XK superfamily. We propose that CDI systems exploit the inherent sequence variability and active-site plasticity of PD(D/E)XK nucleases to generate toxin diversity. These findings raise the possibility that many other uncharacterized CDI toxins may belong to the PD(D/E)XK superfamily.
Keywords: Burkholderia pseudomallei; crystal structure; protein complex; ribonuclease; small-angle x-ray scattering (SAXS); tRNase; toxin; toxin/immunity complexes.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures








References
-
- Makhov A. M., Hannah J. H., Brennan M. J., Trus B. L., Kocsis E., Conway J. F., Wingfield P. T., Simon M. N., and Steven A. C. (1994) Filamentous hemagglutinin of Bordetella pertussis. A bacterial adhesin formed as a 50-nm monomeric rigid rod based on a 19-residue repeat motif rich in β strands and turns. J. Mol. Biol. 241, 110–124 - PubMed
-
- Kajava A. V., Cheng N., Cleaver R., Kessel M., Simon M. N., Willery E., Jacob-Dubuisson F., Locht C., and Steven A. C. (2001) β-Helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol. Microbiol. 42, 279–292 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

