Pharmacophore modeling and in silico toxicity assessment of potential anticancer agents from African medicinal plants
- PMID: 27445461
- PMCID: PMC4938243
- DOI: 10.2147/DDDT.S108118
Pharmacophore modeling and in silico toxicity assessment of potential anticancer agents from African medicinal plants
Abstract
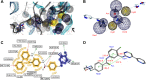
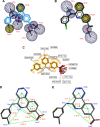
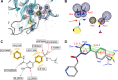
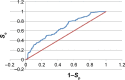
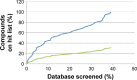
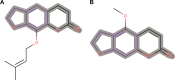
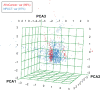
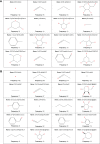
Molecular modeling has been employed in the search for lead compounds of chemotherapy to fight cancer. In this study, pharmacophore models have been generated and validated for use in virtual screening protocols for eight known anticancer drug targets, including tyrosine kinase, protein kinase B β, cyclin-dependent kinase, protein farnesyltransferase, human protein kinase, glycogen synthase kinase, and indoleamine 2,3-dioxygenase 1. Pharmacophore models were validated through receiver operating characteristic and Güner-Henry scoring methods, indicating that several of the models generated could be useful for the identification of potential anticancer agents from natural product databases. The validated pharmacophore models were used as three-dimensional search queries for virtual screening of the newly developed AfroCancer database (~400 compounds from African medicinal plants), along with the Naturally Occurring Plant-based Anticancer Compound-Activity-Target dataset (comprising ~1,500 published naturally occurring plant-based compounds from around the world). Additionally, an in silico assessment of toxicity of the two datasets was carried out by the use of 88 toxicity end points predicted by the Lhasa's expert knowledge-based system (Derek), showing that only an insignificant proportion of the promising anticancer agents would be likely showing high toxicity profiles. A diversity study of the two datasets, carried out using the analysis of principal components from the most important physicochemical properties often used to access drug-likeness of compound datasets, showed that the two datasets do not occupy the same chemical space.
Keywords: anticancer; medicinal plants; natural products; pharmacophore; toxicity; virtual screening.
Figures



References
-
- Gibbs JB. Mechanism-based target identification and drug discovery in cancer research. Science. 2000;287(5460):1969–1973. - PubMed
-
- WHO Media Centre [webpage on the Internet] Fast sheet Nº 297. 2013. [Accessed February 4, 2014]. Available from: http://www.who.int/mediacentre/factsheets/fs297/en/index.html.
-
- [webpage on the Internet] Globocan Fast Stats. 2008. [Accessed August 4, 2013]. Available from: http://www.iarc.fr/en/media-centre/iarcnews/2010/globocan2008.php.
-
- Newman DJ, Cragg GM, Snader KM. The influence of natural products upon drug discovery. Nat Prod Rep. 2000;17(3):215–234. - PubMed
-
- Hostettmann K, Terreaux C. Search for new lead compounds from higher plants. Chimia. 2000;54(11):652–657.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

