Hypothesis and Theory: Revisiting Views on the Co-evolution of the Melanocortin Receptors and the Accessory Proteins, MRAP1 and MRAP2
- PMID: 27445982
- PMCID: PMC4923161
- DOI: 10.3389/fendo.2016.00079
Hypothesis and Theory: Revisiting Views on the Co-evolution of the Melanocortin Receptors and the Accessory Proteins, MRAP1 and MRAP2
Abstract
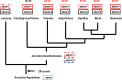
The evolution of the melanocortin receptors (MCRs) is closely associated with the evolution of the melanocortin-2 receptor accessory proteins (MRAPs). Recent annotation of the elephant shark genome project revealed the sequence of a putative MRAP1 ortholog. The presence of this sequence in the genome of a cartilaginous fish raises the possibility that the mrap1 and mrap2 genes in the genomes of gnathostome vertebrates were the result of the chordate 2R genome duplication event. The presence of a putative MRAP1 ortholog in a cartilaginous fish genome is perplexing. Recent studies on melanocortin-2 receptor (MC2R) in the genomes of the elephant shark and the Japanese stingray indicate that these MC2R orthologs can be functionally expressed in CHO cells without co-expression of an exogenous mrap1 cDNA. The novel ligand selectivity of these cartilaginous fish MC2R orthologs is discussed. Finally, the origin of the mc2r and mc5r genes is reevaluated. The distinctive primary sequence conservation of MC2R and MC5R is discussed in light of the physiological roles of these two MCR paralogs.
Keywords: MC2R; MC5R; MRAP1; MRAP2; evolution; melanocortin receptors.
Figures



Similar articles
-
Evaluating the interactions between red stingray (Dasyatis akajei) melanocortin receptors and elephant shark (Callorhinchus milii) MRAP1 and MRAP2 following stimulation with either stingray ACTH(1-24) or stingray Des-Acetyl-αMSH: A pharmacological study in Chinese Hamster Ovary cells.Gen Comp Endocrinol. 2018 Sep 1;265:133-140. doi: 10.1016/j.ygcen.2018.02.018. Epub 2018 Mar 7. Gen Comp Endocrinol. 2018. PMID: 29524525
-
Elephant shark melanocortin receptors: Novel interactions with MRAP1 and implication for the HPI axis.Gen Comp Endocrinol. 2019 Feb 1;272:42-51. doi: 10.1016/j.ygcen.2018.11.009. Epub 2018 Nov 20. Gen Comp Endocrinol. 2019. PMID: 30468718
-
Characterization of melanocortin receptors from stingray Dasyatis akajei, a cartilaginous fish.Gen Comp Endocrinol. 2016 Jun 1;232:115-24. doi: 10.1016/j.ygcen.2016.03.030. Epub 2016 Mar 25. Gen Comp Endocrinol. 2016. PMID: 27021018
-
60 YEARS OF POMC: Melanocortin receptors: evolution of ligand selectivity for melanocortin peptides.J Mol Endocrinol. 2016 May;56(4):T119-33. doi: 10.1530/JME-15-0292. Epub 2016 Jan 20. J Mol Endocrinol. 2016. PMID: 26792827 Review.
-
Views on the co-evolution of the melanocortin-2 receptor, MRAPs, and the hypothalamus/pituitary/adrenal-interrenal axis.Mol Cell Endocrinol. 2015 Jun 15;408:12-22. doi: 10.1016/j.mce.2014.12.022. Epub 2015 Jan 5. Mol Cell Endocrinol. 2015. PMID: 25573240 Review.
Cited by
-
Ligands for Melanocortin Receptors: Beyond Melanocyte-Stimulating Hormones and Adrenocorticotropin.Biomolecules. 2022 Oct 1;12(10):1407. doi: 10.3390/biom12101407. Biomolecules. 2022. PMID: 36291616 Free PMC article. Review.
-
Structure/Function Studies on the Activation Motif of Two Non-Mammalian Mrap1 Orthologs, and Observations on the Phylogeny of Mrap1, Including a Novel Characterization of an Mrap1 from the Chondrostean Fish, Polyodon spathula.Biomolecules. 2022 Nov 12;12(11):1681. doi: 10.3390/biom12111681. Biomolecules. 2022. PMID: 36421695 Free PMC article.
-
Identifying Common Features in the Activation of Melanocortin-2 Receptors: Studies on the Xenopus tropicalis Melanocortin-2 Receptor.Int J Mol Sci. 2019 Aug 26;20(17):4166. doi: 10.3390/ijms20174166. Int J Mol Sci. 2019. PMID: 31454910 Free PMC article.
-
Melanocortin Receptor 4 (MC4R) Signaling System in Nile Tilapia.Int J Mol Sci. 2020 Sep 24;21(19):7036. doi: 10.3390/ijms21197036. Int J Mol Sci. 2020. PMID: 32987823 Free PMC article.
-
Regulation of Melanocortin-3 and -4 Receptors by Isoforms of Melanocortin-2 Receptor Accessory Protein 1 and 2.Biomolecules. 2022 Feb 2;12(2):244. doi: 10.3390/biom12020244. Biomolecules. 2022. PMID: 35204745 Free PMC article.
References
-
- Vastermark A, Schiöth HB. The early origin of melanocortin receptors, agouti-related peptide, agouti signaling peptide, and melanocortin receptor-accessory proteins, with emphasis on pufferfishes, elephant shark, lampreys, and amphioxus. Eur J Pharmacol (2011) 660:61–9.10.1016/j.ejphar.2010.10.106 - DOI - PubMed
-
- Holland PW, Garcia-Fernandez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl (1994):125–33. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

