Gibberellin-Stimulation of Rhizome Elongation and Differential GA-Responsive Proteomic Changes in Two Grass Species
- PMID: 27446135
- PMCID: PMC4917561
- DOI: 10.3389/fpls.2016.00905
Gibberellin-Stimulation of Rhizome Elongation and Differential GA-Responsive Proteomic Changes in Two Grass Species
Abstract
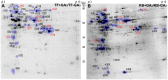
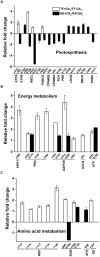
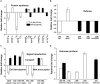
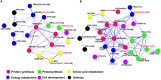
Rapid and extensive rhizome development is a desirable trait for perennial grass growth and adaptation to environmental stresses. The objective of this study was to determine proteomic changes and associated metabolic pathways of gibberellin (GA) -regulation of rhizome elongation in two perennial grass species differing in rhizome development. Plants of a short-rhizome bunch-type tall fescue (TF; Festuca arundinacea; 'BR') and an extensive rhizomatous Kentucky bluegrass (KB; Poa pratensis; 'Baron') were treated with 10 μM GA3 in hydroponic culture in growth chambers. The average rhizome length in KB was significantly longer than that in TF regardless of GA3 treatment, and increased significantly with GA3 treatment, to a greater extent than that in TF. Comparative proteomic analysis using two-dimensional electrophoresis and mass spectrometry was performed to further investigate proteins and associated metabolic pathways imparting increased rhizome elongation by GA. A total of 37 and 38 differentially expressed proteins in response to GA3 treatment were identified in TF and KB plants, respectively, which were mainly involved in photosynthesis, energy and amino acid metabolism, protein synthesis, defense and cell development processes. Accelerated rhizome elongation in KB by GA could be mainly associated with the increased abundance of proteins involved in energy metabolism (glyceraldehyde-3-phosphate dehydrogenase, fructose-bisphosphate aldolase, and ATP synthase), amino acid metabolism (S-adenosylmethionine and adenosylhomocysteinase), protein synthesis (HSP90, elongation factor Tu and eukaryotic translation initiation factor 5A), cell-wall development (cell dividion cycle protein, alpha tubulin-2A and actin), and signal transduction (calreticulin). These proteins could be used as candidate proteins for further analysis of molecular mechanisms controlling rhizome growth.
Keywords: Kentucky bluegrass; gibberellic acid; hydroponics; plant proteomics; tall fescue.
Figures






References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

