Construction of Global Acyl Lipid Metabolic Map by Comparative Genomics and Subcellular Localization Analysis in the Red Alga Cyanidioschyzon merolae
- PMID: 27446184
- PMCID: PMC4928187
- DOI: 10.3389/fpls.2016.00958
Construction of Global Acyl Lipid Metabolic Map by Comparative Genomics and Subcellular Localization Analysis in the Red Alga Cyanidioschyzon merolae
Abstract
Pathways of lipid metabolism have been established in land plants, such as Arabidopsis thaliana, but the information on exact pathways is still under study in microalgae. In contrast with Chlamydomonas reinhardtii, which is currently studied extensively, the pathway information in red algae is still in the state in which enzymes and pathways are estimated by analogy with the knowledge in plants. Here we attempt to construct the entire acyl lipid metabolic pathways in a model red alga, Cyanidioschyzon merolae, as an initial basis for future genetic and biochemical studies, by exploiting comparative genomics and localization analysis. First, the data of whole genome clustering by Gclust were used to identify 121 acyl lipid-related enzymes. Then, the localization of 113 of these enzymes was analyzed by GFP-based techniques. We found that most of the predictions on the subcellular localization by existing tools gave erroneous results, probably because these tools had been tuned for plants or green algae. The experimental data in the present study as well as the data reported before in our laboratory will constitute a good training set for tuning these tools. The lipid metabolic map thus constructed show that the lipid metabolic pathways in the red alga are essentially similar to those in A. thaliana, except that the number of enzymes catalyzing individual reactions is quite limited. The absence of fatty acid desaturation to produce oleic and linoleic acids within the plastid, however, highlights the central importance of desaturation and acyl editing in the endoplasmic reticulum, for the synthesis of plastid lipids as well as other cellular lipids. Additionally, some notable characteristics of lipid metabolism in C. merolae were found. For example, phosphatidylcholine is synthesized by the methylation of phosphatidylethanolamine as in yeasts. It is possible that a single 3-ketoacyl-acyl carrier protein synthase is involved in the condensation reactions of fatty acid synthesis in the plastid. We will also discuss on the redundant β-oxidation enzymes, which are characteristic to red algae.
Keywords: Cyanidioschyzon merolae; comparative genomics; lipid metabolism; red alga; subcellular localization.
Figures
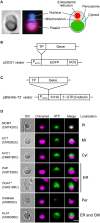

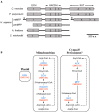
References
-
- Clough R. C., Matthis A. L., Barnum S. R., Jaworski J. G. (1992). Purification and characterization of 3-ketoacyl-acyl carrier protein synthase III from spinach. A condensing enzyme utilizing acetyl-coenzyme A to initiate fatty acid synthesis. J. Biol. Chem. 267, 20992–20998. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

