Genetic lineage tracing defines myofibroblast origin and function in the injured heart
- PMID: 27447449
- PMCID: PMC5512625
- DOI: 10.1038/ncomms12260
Genetic lineage tracing defines myofibroblast origin and function in the injured heart
Abstract
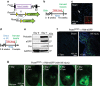
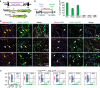
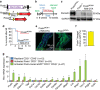
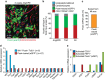
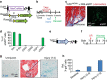
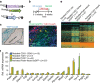
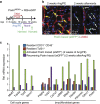
Cardiac fibroblasts convert to myofibroblasts with injury to mediate healing after acute myocardial infarction (MI) and to mediate long-standing fibrosis with chronic disease. Myofibroblasts remain a poorly defined cell type in terms of their origins and functional effects in vivo. Here we generate Postn (periostin) gene-targeted mice containing a tamoxifen-inducible Cre for cellular lineage-tracing analysis. This Postn allele identifies essentially all myofibroblasts within the heart and multiple other tissues. Lineage tracing with four additional Cre-expressing mouse lines shows that periostin-expressing myofibroblasts in the heart derive from tissue-resident fibroblasts of the Tcf21 lineage, but not endothelial, immune/myeloid or smooth muscle cells. Deletion of periostin(+) myofibroblasts reduces collagen production and scar formation after MI. Periostin-traced myofibroblasts also revert back to a less-activated state upon injury resolution. Our results define the myofibroblast as a periostin-expressing cell type necessary for adaptive healing and fibrosis in the heart, which arises from Tcf21(+) tissue-resident fibroblasts.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Ertl G. & Frantz S. Healing after myocardial infarction. Cardiovasc. Res. 66, 22–32 (2005). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

