La Autoantigen Induces Ribosome Binding Protein 1 (RRBP1) Expression through Internal Ribosome Entry Site (IRES)-Mediated Translation during Cellular Stress Condition
- PMID: 27447629
- PMCID: PMC4964545
- DOI: 10.3390/ijms17071174
La Autoantigen Induces Ribosome Binding Protein 1 (RRBP1) Expression through Internal Ribosome Entry Site (IRES)-Mediated Translation during Cellular Stress Condition
Abstract
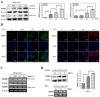
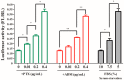
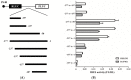
The function of ribosome binding protein 1 (RRBP1) is regulating the transportation and secretion of some intracellular proteins in mammalian cells. Transcription of RRBP1 is induced by various cytokines. However, few studies focused on the process of RRPB1 mRNA translation. The RRBP1 mRNA has a long 5' untranslated region that potentially formed a stable secondary structure. In this study, we show that the 5' UTR of RRBP1 mRNA contains an internal ribosome entry site (IRES). Moreover, the RRBP1 expression is induced by chemotherapeutic drug paclitaxel or adriamycin in human hepatocellular carcinoma cells and accompanied with the increased expression of La autoantigen (La), which binds to RRBP1 IRES element and facilitates translation initiation. Interestingly, we found IRES-mediated RRBP1 translation is also activated during serum-starvation condition which can induce cytoplasmic localization of La. After mapping the entire RRBP1 5' UTR, we determine the core IRES activity is located between nt-237 and -58. Furthermore, two apical GARR loops within the functional RRBP1 IRES elements may be important for La binding. These results strongly suggest an important role for IRES-dependent translation of RRBP1 mRNA in hepatocellular carcinoma cells during cellular stress conditions.
Keywords: La autoantigen (La); adriamycin (ADM); paclitaxel (PTX); ribosome binding protein 1 internal ribosome entry site (RRBP1 IRES); serum-starvation.
Figures





References
-
- Arguelles S., Camandola S., Cutler R.G., Ayala A., Mattson M.P. Elongation factor 2 diphthamide is critical for translation of two IRES-dependent protein targets, XIAP and FGF2, under oxidative stress conditions. Free Radic. Biol. Med. 2014;67:131–138. doi: 10.1016/j.freeradbiomed.2013.10.015. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

