Inference for Stochastic Chemical Kinetics Using Moment Equations and System Size Expansion
- PMID: 27447730
- PMCID: PMC4957800
- DOI: 10.1371/journal.pcbi.1005030
Inference for Stochastic Chemical Kinetics Using Moment Equations and System Size Expansion
Abstract
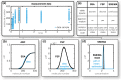
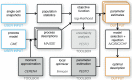
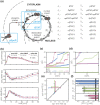
Quantitative mechanistic models are valuable tools for disentangling biochemical pathways and for achieving a comprehensive understanding of biological systems. However, to be quantitative the parameters of these models have to be estimated from experimental data. In the presence of significant stochastic fluctuations this is a challenging task as stochastic simulations are usually too time-consuming and a macroscopic description using reaction rate equations (RREs) is no longer accurate. In this manuscript, we therefore consider moment-closure approximation (MA) and the system size expansion (SSE), which approximate the statistical moments of stochastic processes and tend to be more precise than macroscopic descriptions. We introduce gradient-based parameter optimization methods and uncertainty analysis methods for MA and SSE. Efficiency and reliability of the methods are assessed using simulation examples as well as by an application to data for Epo-induced JAK/STAT signaling. The application revealed that even if merely population-average data are available, MA and SSE improve parameter identifiability in comparison to RRE. Furthermore, the simulation examples revealed that the resulting estimates are more reliable for an intermediate volume regime. In this regime the estimation error is reduced and we propose methods to determine the regime boundaries. These results illustrate that inference using MA and SSE is feasible and possesses a high sensitivity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Gillespie DT. A rigorous derivation of the chemical master equation. Physica A; 1992;188(1):404–425. 10.1016/0378-4371(92)90283-V - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

