Metabolomic Profiling of Human Urine as a Screen for Multiple Inborn Errors of Metabolism
- PMID: 27448163
- PMCID: PMC5314726
- DOI: 10.1089/gtmb.2015.0291
Metabolomic Profiling of Human Urine as a Screen for Multiple Inborn Errors of Metabolism
Abstract
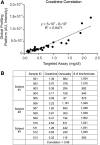
Aims: We wished to determine the efficacy of using urine as an analyte to screen for a broad range of metabolic products associated with multiple different types of inborn errors of metabolism (IEMs), using an automated mass spectrometry-based assay. Urine was compared with plasma samples from a similar cohort analyzed using the same assay. Specimens were analyzed using two different commonly utilized urine normalization methods based on creatinine and osmolality, respectively.
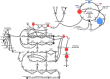
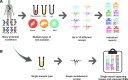
Methods: Biochemical profiles for each sample (from both affected and unaffected subjects) were obtained using a mass spectrometry-based platform and population-based statistical analyses.
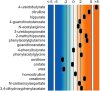
Results: We identified over 1200 biochemicals from among 100 clinical urine samples and identified clear biochemical signatures for 16 of 18 IEM diseases tested. The two diseases that did not result in clear signatures, X-linked creatine transporter deficiency and ornithine transcarbamylase deficiency, were from individuals under treatment, which masked biomarker signatures. Overall the process variability and coefficient of variation for isolating and identifying biochemicals by running technical replicates of each urine sample was 10%.
Conclusions: A single urine sample analyzed with our integrated metabolomic platform can identify signatures of IEMs that are traditionally identified using many different assays and multiple sample types. Creatinine and osmolality-normalized data were robust to the detection of the disorders and samples tested here.
Conflict of interest statement
A.K., A.E., J.W., K.B., and L.M. are employees of Metabolon, Inc. and, as such, have affiliations with or financial involvement with Metabolon, Inc. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the article apart from those disclosed. M.J.M., V.R.S., Q.S., and S.H.E. are employees of Baylor College of Medicine, which has a partnership with Baylor Miraca Genetics Laboratories that derives revenue from clinical testing.
Figures


References
-
- Armstrong LE, Johnson EC, Munoz CX, et al. (2013) Evaluation of Uosm:Posm ratio as a hydration biomarker in free-living, healthy young women. Eur J Clin Nutr 67:934–938 - PubMed
-
- Atwal PS, Donti TR, Cardon AL, et al. (2015) Aromatic l-amino acid decarboxylase deficiency diagnosed by clinical metabolomic profiling of plasma. Mol Genet Metab 115:91–94 - PubMed
-
- Campeau PM, Scriver CR, Mitchell JJ. (2008) A 25-year longitudinal analysis of treatment efficacy in inborn errors of metabolism. Mol Genet Metab 95:11–16 - PubMed
-
- Curcio R, Stettler H, Suter PM, et al. (2015) Reference intervals for 24 laboratory parameters determined in 24-hour urine collections. Clin Chem Lab Med 54:105–116 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

